Background
Recombinant proteins play a key role in many applications, including industrial biocatalysis and therapeutics. Despite recent advances in computational protein structure prediction, reducing protein solubility and aggregation resistance remains a design challenge. Identifying aggregation-prone regions is crucial for understanding misfolding diseases or designing efficient protein technologies, and therefore has enormous socioeconomic impact.

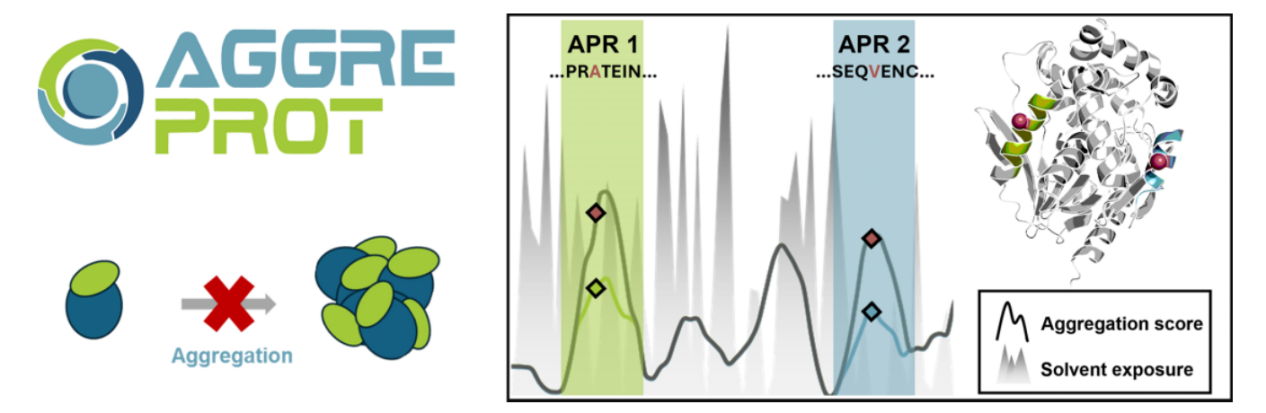
On May 27, 2024, Danish researchers published an article titled "AggreProt: a web server for predicting and engineering aggregation-prone regions in proteins" in Nucleic Acids Research. The article introduces AggreProt, a web server for predicting and engineering aggregation-prone regions in proteins. It automatically leverages an ensemble of deep neural networks to predict aggregation-prone regions (APRs) in protein sequences. Trained on experimentally evaluated hexapeptides, AggreProt compares to or outperforms state-of-the-art algorithms on two independent benchmark datasets. The server provides an intuitive interface with interactive sequence and structure viewers, providing per-residue aggregation profiles, as well as information on solvent accessibility and transmembrane propensity for comprehensive analysis. The researchers demonstrated AggreProt's effectiveness in predicting diverse aggregation behaviors of proteins across several use cases, highlighting its potential for guiding protein engineering strategies to reduce aggregation propensity and improve solubility. The web server is freely available at https://loschmidt.chemi.muni.cz/aggreprot/.
Current issues with protein expression
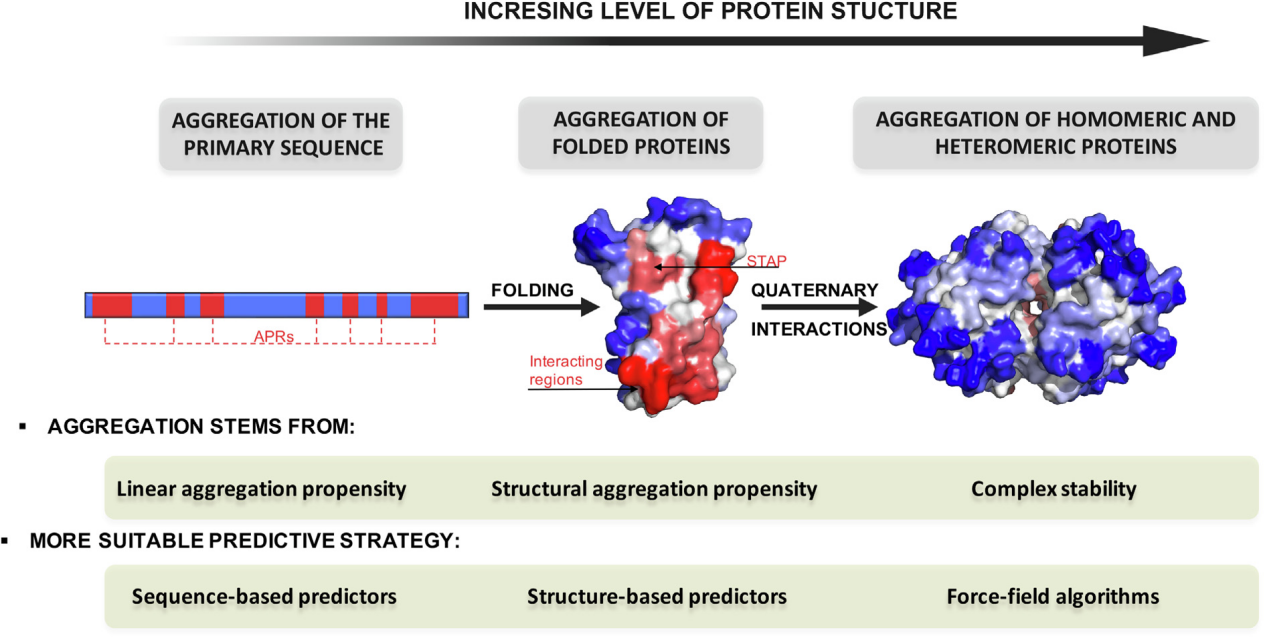
Recombinant proteins play a key role in numerous applications, including industrial biocatalysis and therapeutics. However, inclusion body formation, low purified protein yields, and aggregation/precipitation are common problems in protein expression. Protein folding is primarily driven by the burial of hydrophobic residues; exposure of these residues can lead to unnatural self-association, misfolding, and ultimately aggregation. The formation of these misfolded aggregates can be triggered by a variety of factors and is associated with serious diseases such as Alzheimer's and Parkinson's diseases. Among these aggregates, amyloid proteins represent a unique class characterized by a highly organized two-dimensional structure. Amyloid proteins are formed from stacked repeating units of protein molecules, stabilized by a network of hydrogen bonds within their cross-β-sheet structure; however, these molecules typically adopt distinct conformations. They share a common structural core, which is believed to be a key driver of amyloid formation and crucial for its stability. Therefore, these regions (APRs) are ideal targets for designing mutations that can reduce aggregation propensity and thereby improve protein solubility.
Several algorithms have been designed to address the aggregation problem. Depending on the type of input data they accept, these algorithms are categorized as sequence or structure prediction. Algorithms from these two categories have greatly advanced our understanding of protein aggregation and solubility at the molecular level and are frequently used to identify APRs in proteins, with varying degrees of success.
(Data source: Santos J, et al. Comput Struct Biotechnol J. 2020)
In the last few years, a third generation of predictions based on machine learning has also emerged, such as the Support Vector Machine in the Amyloid Predictor, the Random Forest Classifier in RF-Amyloid and Amylogram, and many others including ANiPP, FishAmy-loid, or CORDAX.

(Data source: Prabakaran R, et al. J Mol Biol. 2021)
Features and advantages of AggreProt
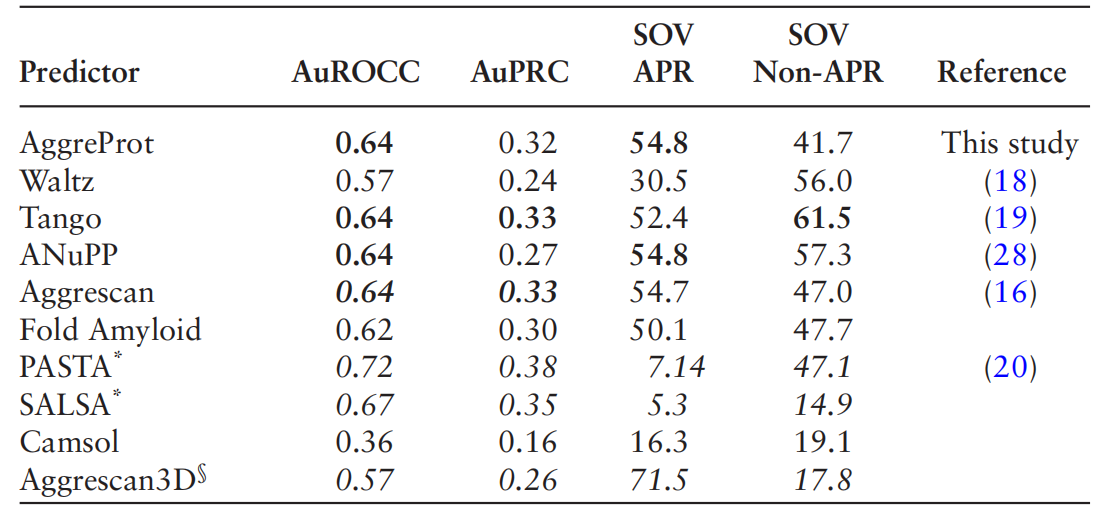
AggreProt can predict aggregation propensity of entire protein sequences, not just individual hexapeptide fragments, and achieves similar or better performance than other state-of-the-art methods based on sequence or machine learning for residue-level or SOV validation.
AggreProtweb server usage process
The server combines its dedicated amyloid aggregation propensity predictor with transmembrane (TM) propensity and solvent accessible surface area (SASA) calculations to provide structural context for the analyzed protein sequences.
Data Input: Users enter protein sequences in FASTA format. The server quickly checks the sequence integrity, including the presence of headers and sequences. The server allows up to three different protein sequences to be entered simultaneously. Users can upload structure files associated with the input sequences (both PDB and mmCIF formats are accepted). If the user cannot provide a structure file, AggreProt offers the option of retrieving the structure from AlphaFoldDB.

Result output: After the calculation is completed, the job status will change to "Completed" and the results will be displayed in graphical form, including an aligned profile graph and a sequence display graph. Users can change the sensitivity and specificity of the aggregation tendency by adjusting the threshold. The aggregation tendency curve is represented by a semi-transparent solid color, and the TM tendency and SASA are represented by dots and dashed lines, respectively. In this part of the visualization tool (tendency graph), hovering the mouse over any sequence position will display additional information about the protein residues and the individual prediction values calculated for each tendency. An interactive sequence and structure viewer is provided, allowing users to compare the profile graphs of multiple proteins. If the user provides a three-dimensional structure of the protein or obtains the structure from the database, AggreProt will provide an interactive three-dimensional view.

Case Verification
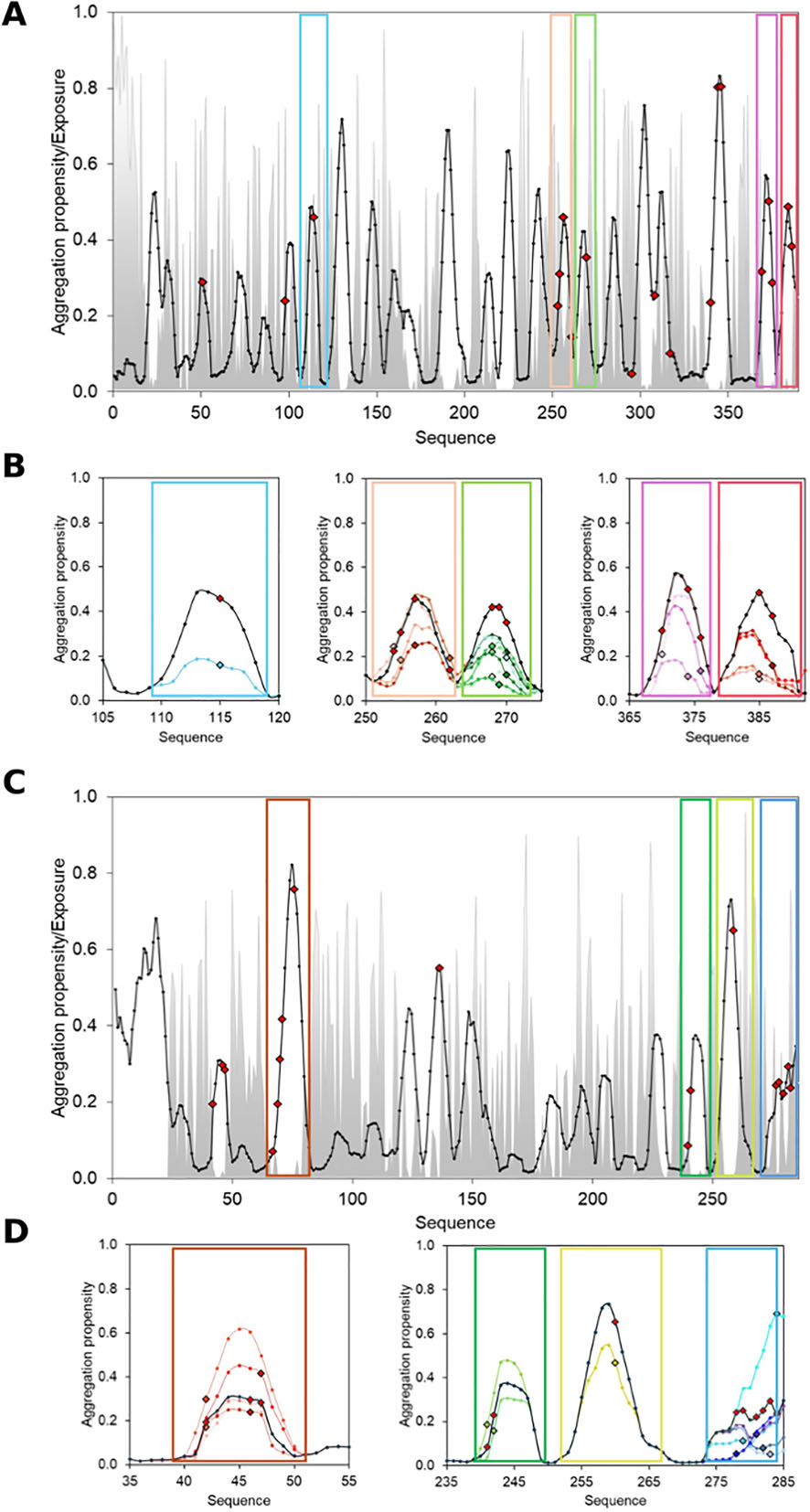
AggreProt was used to identify aggregation-prone regions (APRs) in HLDLinB. Based on AggreProt's predictions, researchers designed a series of mutations aimed at reducing HLDLinB's aggregation propensity and improving its solubility. Experimental evaluation of the designed HLDLinB mutations demonstrated that AggreProt correctly identified mutations that reduced aggregation propensity and increased soluble protein production. AggreProt also demonstrated a superior ability to detect APRs in LinB compared to other predictors.
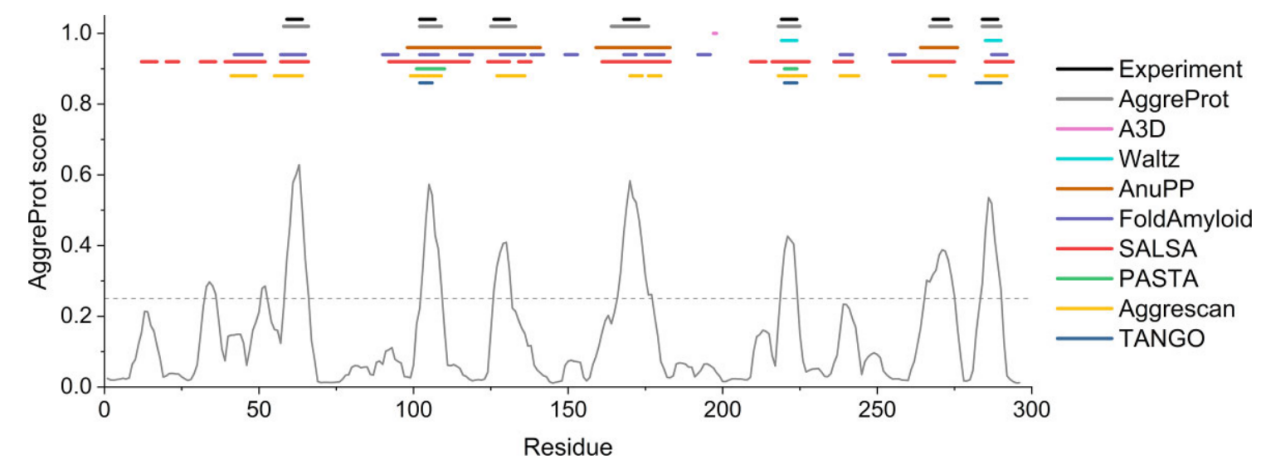
Using deep mutational scanning data from the SoluProtMutDB database, we analyzed type III polyketide synthases and TEM β-lactamases. We found that many solubility-enhancing mutations corresponded to APRs predicted by AggreProt. The observed effects of the mutations were consistent with those predicted by AggreProt in multiple cases, confirming the effectiveness of AggreProt. While AggreProt is able to predict the effects of mutations on aggregation propensity well for surface-exposed APRs, the prediction of effects for buried APRs is more complex. If a mutation increases local hydrophobicity, AggreProt may predict an increase in aggregation propensity, contrary to its prediction for exposed APRs. The accuracy of AggreProt's predictions is closely related to the nature of the mutation, including its type, location (surface or buried), and effect on protein structure and solvent accessibility.
Summary and Outlook
AggreProt is a deep neural network-based web server that predicts aggregation-prone regions (APRs) in protein sequences. AggreProt performs as well as or better than existing state-of-the-art algorithms on two independent benchmark datasets. AggreProt provides an intuitive interface that includes sequence and structure visualization, as well as analysis of aggregation propensity, transmembrane propensity, and solvent accessibility.
AggreProt performs well in predicting the effects of protein mutations on aggregation behavior and can provide guidance for protein engineering. Since the ultimate goal of reducing the aggregation tendency of a protein can only be achieved by modifying its sequence, researchers will enhance the user interface in the future: providing a "design" panel that allows users to plan and predict the results of protein engineering strategies. Allowing users to fine-tune the boundaries of APRs and define custom regions to implement engineering strategies more precisely. Implementing specific mutation strategies for APRs, such as substitution of gatekeeper residues and saturation mutation of exposed residues. Providing more customization options allows users to select and combine different mutation strategies. Displaying ideal multi-mutation sequences evaluated by AggreProt for users to review and use. By continuing to improve the performance of AggreProt, it will become a more powerful protein engineering tool.
