Background
The complementarity-determining regions (CDRs) of antibodies are the most diverse regions in terms of sequence and structural characteristics and play a critical role in antibody recognition and binding immune responses. Over the past few decades, several sequence-based coding schemes have been introduced, such as Kabat, Chothia, AbM, and IMGT. However, the existence of multiple coding schemes has led to potential confusion and a lack of comprehensive evaluation of these schemes.

On December 4, 2024, an article titled "50 Years of Antibody Numbering Schemes: A Statistical and Structural Evaluation Reveals Key Differences and Limitations" was published in Antibodies (Basel). The study used statistical analysis to quantify the diversity of CDR numbering schemes . The study revealed regional sequence and structural conservation in antibody sequence databases, while also highlighting the differences caused by different numbering schemes. These insights provide valuable guidance for the precise description of antibody CDRs and the strategic design of antibody libraries, and have practical significance for the development of innovative antibody-based therapeutics and diagnostics.

Kabat numbering scheme
Compared to the framework, CDR residues using the Kabat numbering scheme exhibit moderately lower conservation, though some residues are more diverse than others. The average relative entropy values for the entire variable domain, framework region, and CDRs were 1.65, 1.94, and 0.92, respectively. Higher relative entropy values indicate less variation or greater conservation. Relative entropy analysis revealed that CDR regions are less conserved than framework regions.

Analysis of different numbering schemes
Several positions within the annotated CDRs showed remarkable structural conservation. These positions were primarily located at the ends of the CDR loops. Two regions with highly conserved structural properties were also found: one located from L53 to L56 in LCDR2 and the other located from H57 to H65 in HCDR2. These regions aligned with the boundaries designated as CDRs within the Kabat and AbM numbering schemes, but not in the Chothia and IMGT numbering schemes. Structural alignments showed that these two regions are not loop regions, but rather contain a conserved β-sheet and an extended loop structure. Parallel analysis of sequence and structure showed that Kabat and AbM include more conserved residues in their CDR definitions than Chothia and IMGT. Both the Chothia and IMGT schemes appear to underestimate the residues included in LCDR2.

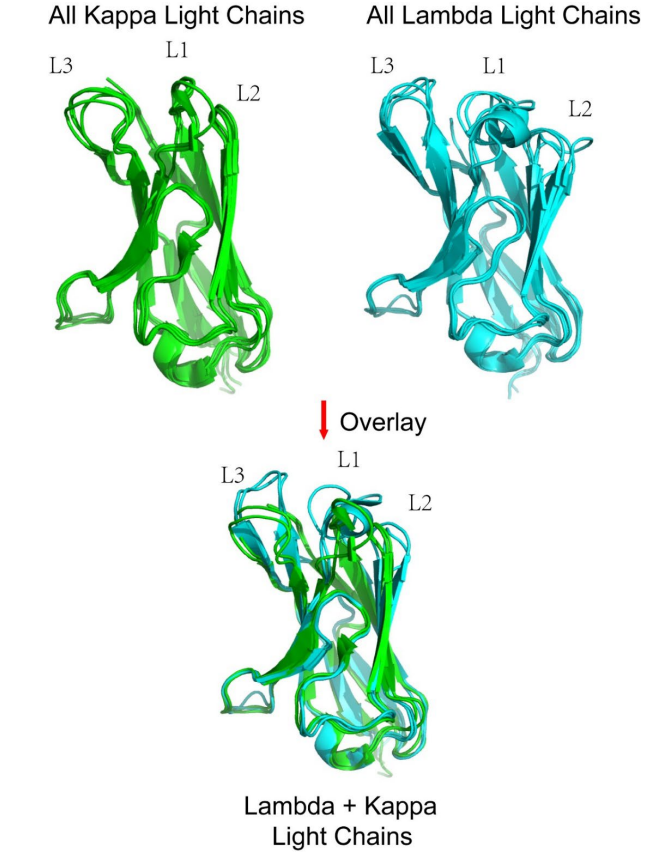
Lambda light chains show significant homology to kappa light chains, but there are significant differences between kappa and lambda light chains.
Amino acid distribution of LCDR1 and HCDR1
A structurally conserved "pivot" point is observed in CDR1, located between L29 and H30 (or H29 in Kabat, AbM, and Chothia), which divides CDR1 into two loops whose side chains dip into a hydrophobic pocket between two β sheets. Significant structural differences are observed between kappa and lambda light chain structures, but within the LCDR1 region, a pivot point is observed in both types, but exhibits different amino acid preferences in the light and heavy chains.
The C-terminal loop within the CDR1 region shows greater sequence diversity than the N-terminus. Other studies have also shown that the C-terminal loop interacts more directly with antigen than the N-terminal loop. These findings collectively suggest that the C-terminal loop may exhibit more CDR-like features than the N-terminal loop.

Distribution of amino acids in the framework and CDRs
Framework Regions: The overall frequency of large aromatic amino acids within the variable domains is roughly consistent with that of the protein as a whole. Gly and Ser frequencies increased significantly, with their occurrence increasing by 80% and 50%, respectively. Valine (Val) and threonine (Thr) are also more common within the β-sheet structure of the framework regions, consistent with their β-sheet orientation. Tryptophan (Trp) is also more common within the framework regions.
CDRs: Tyr is particularly frequent. The presence of the Tyr aromatic ring, combined with the hydroxyl group, promotes various types of interactions with the antigen, including H-bonds and hydrophobic interactions. The results suggest that Tyr is concentrated in regions directly binding to the antigen. Ser and Gly are both common amino acids in CDRs. Gly has a substantial impact on the formation of loop structures rather than contacting the antigen. Gly is significantly enriched in intercalating residues (such as H100A), especially within HCDR3. These amino acids may not directly participate in antigen interactions but may play a supporting role in maintaining loop structure and interacting with solvent molecules.

Impact of humanization
Differences in framework regions: Light chain framework region 1 (LFR-1) and heavy chain framework region 3 (HFR-3) show higher relative entropy values between humans and mice, indicating significant sequence divergence. The amino acid composition of these regions differs in mouse antibodies from that in human antibodies, requiring special attention during humanization to reduce immunogenicity.
Although the overall relative entropy value of the heavy chain framework region 4 (HFR-4) is low, some individual sites (such as H108 and H109) still show relatively high relative entropy values, suggesting that there may be potential humanization hotspots at these sites.
Differences in specific amino acid positions: The L27B (Kabat numbering) or L29 (IMGT numbering) positions exhibit significant differences in amino acid distribution between human and mouse antibodies. In human antibodies, the L27B position is more biased toward polar amino acids, while in mouse antibodies, it is predominantly nonpolar. This difference is an important consideration during humanization.
Differences in CDRs: The "fulcrum" site (L29) in LCDR1 is more inclined to polar amino acids (such as Asn and Asp) in human antibodies, while it is mainly non-polar amino acids in mouse antibodies. This difference may be related to the different ratios of lambda light chains to kappa light chains between humans and mice. The average kappa to lambda light chain ratio in humans is 2:1, while it is lower in mice.

Characterization of antibody sequence conservation levels
Most highly conserved amino acid residues are located in structurally conserved regions, indicating a close correspondence between sequence conservation and structural conservation, which provides important clues for understanding the relationship between antibody function and structure. A remarkably conserved β-sheet, defined by the "RFSGSXSG" pattern, is present, with over 90% conservation within the light chain variable domain. Within HCDR3, amino acid residues at certain positions (such as H105, H106, and H116) are highly conserved, and the conservation of these residues may be related to V(D)J gene rearrangements.

In conclusion
Using studies of amino acid frequencies and relative entropy across various numbering schemes, existing numbering schemes were evaluated from the perspective of sequence conservation and diversity. It was found that no single numbering scheme fully encompasses all of the most variable residues in the CDRs, and that these schemes differ in their inclusion of adjacent framework-like residues. Specifically, within LCDR1, differences between kappa and lambda chains lead to inconsistent numbering around this feature. These findings can help researchers select the most appropriate numbering scheme for their specific research goals. These insights provide valuable guidance for accurately defining antibody CDRs and strategically designing antibody libraries, offering practical implications for the development of innovative antibody-based therapeutics and diagnostics to address and prevent human diseases.
