Background
Rigorous assessment of the developability of antibodies is crucial for optimizing drug candidates before entering clinical studies. Recent advances in tools for predicting protein structure, surface properties, stability, and immunogenicity have streamlined the development of new biologics. However, due to the diversity of complementarity-determining regions (CDRs), particularly CDR3, accurately predicting the effect of single amino acid substitutions on antibody structure remains challenging.
On December 20, 2024, researchers published an article titled "Integrating In Silico and In Vitro Tools for Optimized Antibody Development—Design of Therapeutic Anti-oxMIF Antibodies" in Antibodies (Basel). The study combined computer simulation tools and in vitro evaluation to improve antibodies targeting oxidative macrophage migration inhibitory factor (oxMIF), based on the first-generation anti-oxMIF antibody imalumab. By introducing mutations in the variable regions, the candidate antibody C0083 exhibited reduced hydrophobicity and self-interactions due to the remodeling of its heavy chain CDR3 loop. Despite these structural changes, C0083 retained its target specificity and binding affinity for oxMIF, demonstrating that a small number of carefully selected mutations is sufficient to significantly improve antibody performance.

Antibody molecular characteristics analysis
The Fab fragment of Imalumab forms Fab dimers upon crystallization, and these dimers are stabilized by hydrophobic interactions. The interactions between the two Fabs are driven by H:L98, H:W97, H:Y99, L:Y32, L:F92, and L:W93.

Aggregation and hydrophobicity analysis: Both amino acids H:W97 and L:W93 are located in the corresponding CDR3 regions. CDR3 is considered the most important region for determining antigen specificity, so caution should be exercised when making mutations in these regions as they may significantly disrupt functional antibody binding.
Post-translational modification analysis: Tryptophan and methionine are known to be susceptible to oxidation, posing a threat to the stability of monoclonal antibodies and ultimately affecting their efficacy and safety. Residues H:W97, L:M30, and L:W93 are highly solvent-exposed, making them susceptible to oxidation and therefore selected as candidate sites for mutation.
Immunogenicity Risk Assessment: By analyzing the VH and VL sequences, high affinity peptides were predicted to bind to MHC class II T cells to reduce immune responses. The heavy chain did not display any high-affinity MHC class II epitopes, while two potential core epitopes were identified in the light chain: I29MTYLNWYQ37 (HLA-DRB1*15:01) and F49VASHSQSG57 (HLA-DRB1*01:01 and HLA-DRB1*15:01).
Framework optimization: By comparing the VL and VH domains of imalumab with all human antibodies in AbYsis and the VH3 and Vκ1 sequences in the IMGT germline library, amino acids that are unusual or have hydrophobic properties in the framework regions were identified: P41, S49, R83, and A84 in VH, and D1, Q3, L11, V15, P80, and S83 in VL.

Optimizing mAb variant design
Recombinant mutant antibodies of selected mutants were designed by analyzing increased surface hydrophobicity and aggregation propensity, high-affinity T-cell epitopes, oxidation sites, and several uncommon amino acid residues in the framework and screened using computer modeling.
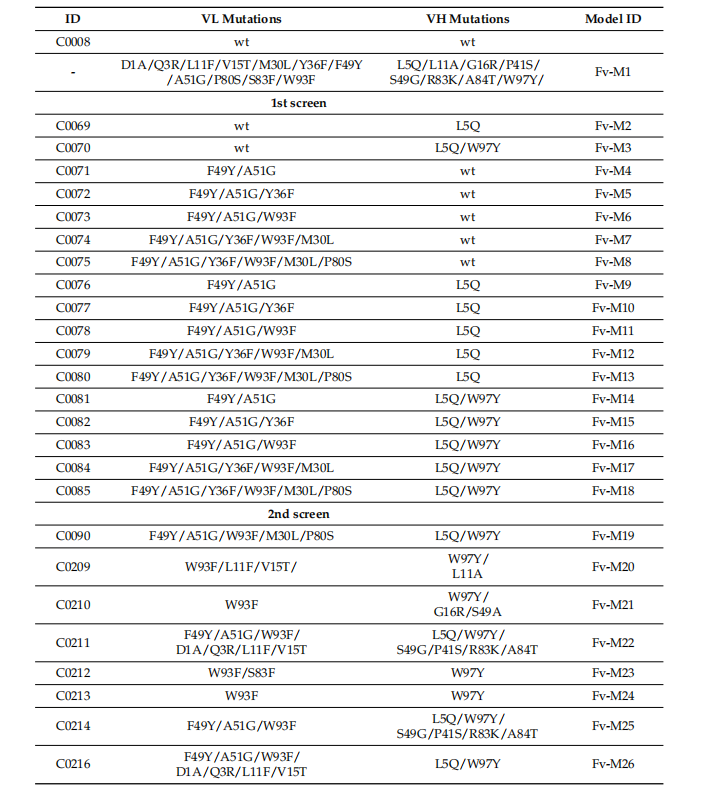
Computer modeling to optimize antibody variants
Modeling of the optimized mAb variants using the AbodyBuilder structure prediction model revealed a significant beneficial effect of the introduced mutations. Compared to imalumab, the hydrophobicity index of L5Q, P41S, and W97Y in the heavy chain (HC) and V15T, F49Y, and W93F in the light chain (LC) decreased. The most hydrophobic amino acids, H:W97 and L:W93, significantly influence the surface hydrophobicity of imalumab, and therefore the introduction of the H:W97Y and L:W93F mutations is expected to reduce the overall hydrophobicity of imalumab.

Characterization of optimized antibodies
The optimized mAb variants all retained their specificity for oxMIF. The introduced mutations did not disrupt the antibody's antigen-binding site, allowing the antibodies to recognize and bind to the target antigen, oxMIF. Variants carrying the L:Y36F mutation (such as C0072 and C0074) exhibited significantly reduced binding affinity for oxMIF and lower thermal stability, indicating that L:Y36F is crucial for proper light chain assembly.
Selected mutations significantly reduced the mAb's surface hydrophobicity and aggregation propensity. In particular, the CDR3 mutations L:W93F and H:W97Y were crucial for reducing self-interactions and aggregation. L:M30 was found to have low sensitivity to oxidation. The introduction of the L:A51G mutation successfully removed a predicted high-affinity T-cell epitope 2, reducing the antibody's immunogenicity risk.

Based on mAb specificity and affinity as assessed by oxMIF, SEC, HIC, AC-SINS, and in silico immunogenicity summary data, C0083 was identified as the most promising mAb variant. It retains the binding properties of imalumab while exhibiting significantly lower hydrophobicity and aggregation potential, as well as reduced immunogenicity risk due to the removal of predicted T-cell epitopes.
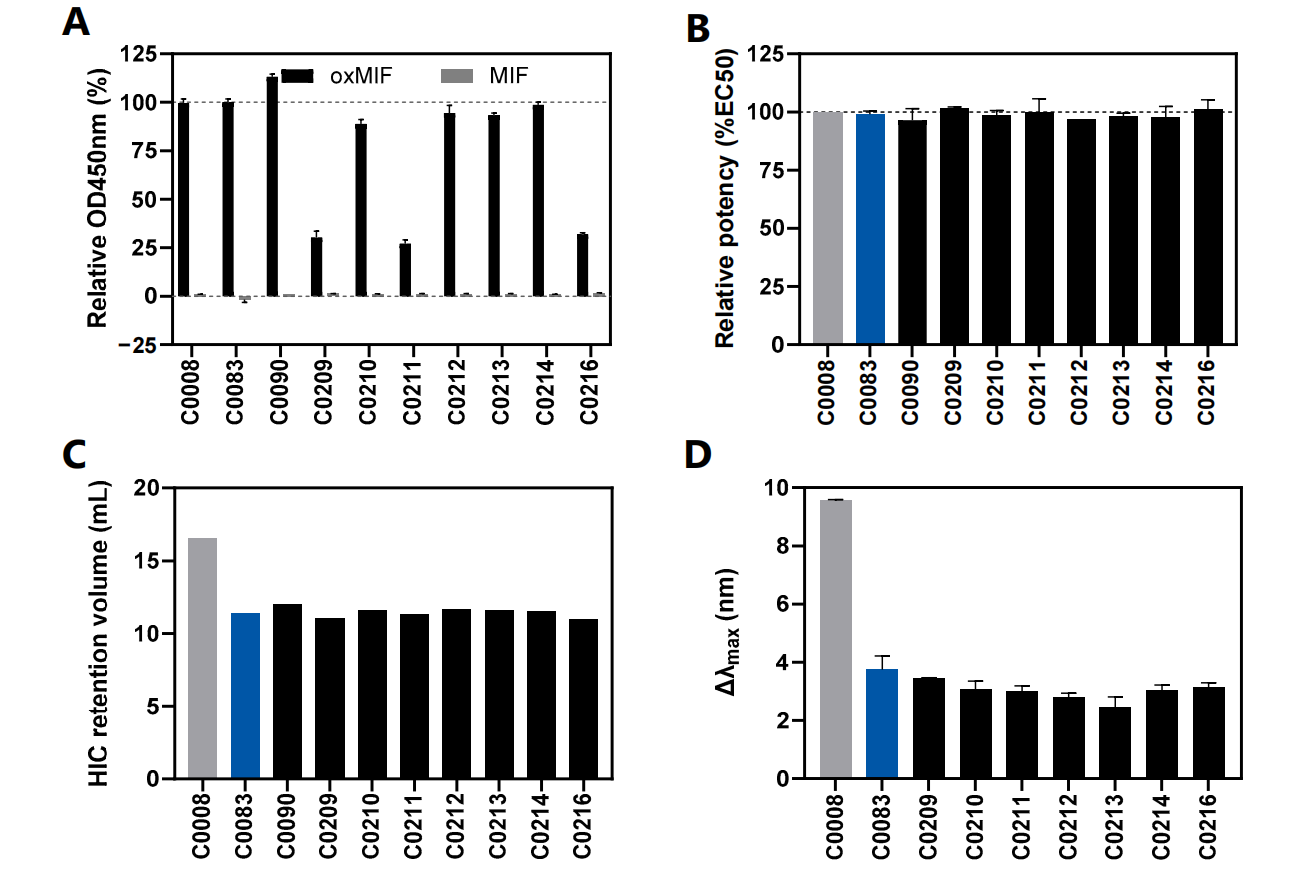
Pharmacokinetic and structural analysis
The tumor half-life of C0083 is prolonged compared to imalumab, and this engineering modification significantly improved the PK and BD profiles of C0083. C0083 is directly related to reduced hydrophobicity and aggregation tendency through targeted mutations in its variable regions.
Crystallization studies of the C0083 Fab revealed a significant rearrangement of the H:CDR3 loop. This rearrangement reduced hydrophobicity and aggregation propensity. Despite these structural changes, C0083 maintained its specificity and high affinity for oxMIF.
However, relying solely on models without biophysical and chemical experimental verification may lead to erroneous conclusions and may miss promising candidate antibodies such as C0083.

In conclusion
Computer simulations and experimental validation identified the surface hydrophobicity and aggregation propensity of imalumab, a first-generation anti-oxMIF antibody, as potential contributors to its unusually short half-life. Introduction of point mutations H:L5Q/W97Y and L:F49Y/A51G/W93F into the variable region of imalumab significantly improved the biophysicochemical properties of the candidate antibody while maintaining high affinity and specificity for oxMIF. C0083, harboring these mutations, was identified as a lead candidate due to its significantly reduced surface hydrophobicity and aggregation potential. Furthermore, it exhibited minimal immunogenicity risk and improved pharmacokinetic properties. Structural analysis of the C0083 Fab revealed that rearrangement of the heavy chain CDR3 loop was a key factor in the observed enhanced biophysicochemical properties. This work demonstrates how molecular defects can be addressed through bioengineering to optimize antibodies, making C0083 and its derivatives promising candidates for the targeted treatment of cancer and inflammatory diseases and advancing them toward clinical validation.
