Background
Protein expression is a critical process in a wide range of biological systems. Escherichia coli, a microbial host widely used in industrial catalysis and healthcare, often presents significant challenges when constructing recombinant expression systems. To maximize the potential of E. coli expression systems, addressing the low or absent yields of certain target proteins is crucial.
On July 20, 2024, researchers from the Key Laboratory of Industrial Biocatalysis, Ministry of Education published an article titled "Strategies to overcome the challenges of low or no expression of heterologous proteins in Escherichia coli" in Biotechnology Advances. The article addresses the major challenges facing exogenous protein expression in Escherichia coli, including protein toxicity, intrinsic effects of gene sequence, and mRNA structure, and proposes feasible solutions. These strategies include specialized methods for managing the expression of toxic proteins, addressing issues related to mRNA structure and codon bias, advanced codon optimization methods that consider multiple factors, and emerging optimization technologies driven by big data and machine learning.

Advantages and current status of heterologous expression of recombinant proteins in Escherichia coli
Protein expression is a fundamental tool in molecular biology and biotechnology, playing an indispensable role in the research, diagnosis, and treatment of various diseases, as well as in industrial bioproduction. Escherichia coli is one of the most widely used hosts for recombinant protein expression, offering advantages such as rapid growth, simple culture conditions, and a wealth of molecular tools for research. Recombinant protein expression in E. coli primarily involves host selection, gene cloning, vector construction, optimization of expression conditions, and verification of expression results.

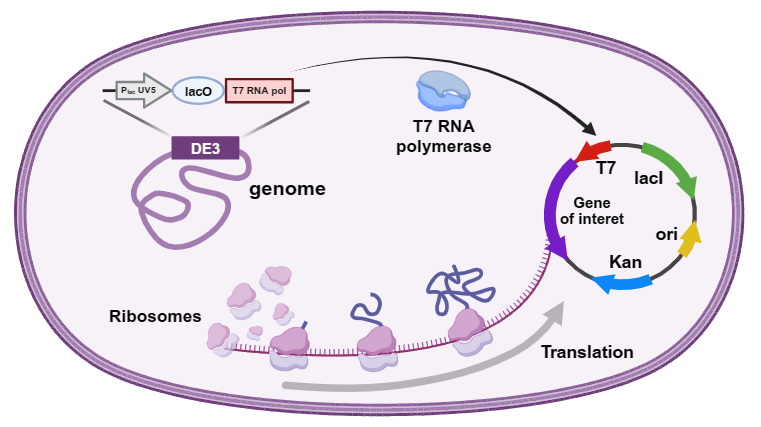
To enhance recombinant protein expression, a variety of plasmid vectors and E. coli strains have been developed, including the pET series of plasmids and the E. coli strain BL21 (DE3) using the T7 expression system. Currently, expression of foreign proteins in E. coli can result in no expression, low expression levels, or expression in inclusion bodies. Existing approaches to optimizing protein expression primarily focus on strategies such as promoter replacement, simple codon optimization, host selection, and optimized culture conditions.
Protein toxicity
Protein toxicity is a common challenge in recombinant protein expression in Escherichia coli. When these proteins disrupt normal host physiological processes, they can cause growth inhibition or even cell death. Common toxic proteins include ribonucleases that cleave mRNA translation start sites, the DNA-binding protein CENP-B, and others that are toxic to the host during the pre-induction growth phase. Some proteins, such as membrane proteins, are only toxic when overexpressed (after induction). Various intracellular synthesis strategies are available for addressing toxic proteins. These strategies primarily focus on minimizing basal expression of toxic proteins during bacterial growth and achieving high expression levels after induction, thereby ensuring sufficient target protein production before host cell death. Numerous strains are currently available for expressing toxic proteins. For example, widely used strains such as BL21(DE3)pLysS and pLysE carry the T7 RNAP inhibitor lysozyme, which reduces the transcriptional strength of the T7 promoter before induction, thereby reducing host toxicity.

In addition to strictly regulating intracellular protein synthesis to mitigate toxicity, another strategy involves expressing toxic proteins and secreting them into the extracellular space to avoid disrupting intracellular growth. This approach typically involves fusing a signal peptide to the N-terminus of the target protein to promote secretory expression of the desired protein. In E. coli, common signal peptides include OmpA, OmpF, PelB, LamB, and PhoA. E. coli has two secretion pathways: type I and type II. The type II secretion pathway is widely used to secrete target proteins into the periplasm. This pathway includes the Sec-dependent pathway, the signal recognition particle (SRP) pathway, and the twin-arginine translocation (TAT) pathway. The Sec-dependent pathway is suitable for proteins that are unfolded in the cytoplasm. The SRP pathway can secrete proteins that are already folded in the cytoplasm. The TAT pathway is more suitable for secretion of complex proteins that require complete intracellular folding or contain disulfide bonds. Numerous proteins have been successfully secreted in E. coli using appropriate secretion pathways. For example, the OmpF signal peptide was used to secrete human interleukin-10 through a seconds-dependent pathway; the signal peptide enhancer B1 (MERACVAV) was used to optimize the PelB and MalE signal peptides to secrete PET hydrolase (IsPETase) from Ideonella sakaiensis through a seconds-dependent pathway; the Dsb signal peptide was used to secrete single-chain Fv (scFv) through the SRP pathway, etc.

Gene sequence determines protein expression
During the process of microbial protein synthesis, a variety of factors can lead to improper protein synthesis, including gene replication, transcription, and translation. The main factor leading to the non-expression of recombinant proteins may be due to differences in gene sequences, which lead to changes in mRNA.
Translational impairment caused by codon bias
Codon bias is considered one of the most critical factors affecting protein expression. During translation, the concentration of charged tRNA corresponding to different codons varies, as does the decoding rate of different codons. Extremely low decoding rates and the absence of charged tRNAs can lead to translational disruptions, resulting in protein expression problems. Factors that may influence codon decoding rates include tRNA modification and activation processes, tRNA diffusion dynamics, and the affinity between codons and anticodons. Ribosomes may slow down or pause at specific locations, such as consecutive proline residues, positively charged amino acids, and SD-like sequences.

The structure of mRNA
When studying the effects of heterologous gene sequences on protein expression in Escherichia coli, the impact of mRNA is crucial. However, directly observing the secondary structure of mRNA prior to ribosome binding remains a significant challenge. Several mRNA structure prediction and mapping methods have been developed, including free energy minimization, suboptimal structure prediction, base pairing probability prediction, parallel analysis of RNA structure (PARS) mapping, and chemical modification detection. Cryo-electron microscopy (cryo-EM) offers the potential to simultaneously explore the 3D structure and conformational dynamics of RNA, leveraging artificial intelligence to predict both secondary and tertiary mRNA structure. Analysis using these mRNA structure determination methods revealed that mRNA secondary structure has the most significant impact on expression levels. Reducing mRNA structural complexity significantly improves expression levels in mutants, and reducing the 5' end structure of the CDS facilitates translation initiation and gene expression. mRNA structure is a fundamental and widespread regulatory factor in gene expression, guiding gene expression in E. coli.

Joint influence of multiple parameters
The expression of recombinant proteins is jointly regulated by codon usage and mRNA structure, and changes in codon usage can affect mRNA structure. Ribosome profiling results show that during the translation initiation phase, ribosomes are densely enriched in a region known as the "ramp sequence." The "ramp hypothesis" suggests that the presence of these codons is intended to slow the rate of translation elongation, thereby reducing the likelihood of ribosome interference with mRNA and preventing ribosome detachment. The "structural hypothesis" proposes that these codons are intended to reduce the structural folding of mRNA at the start site. Studies have found that the impact of codon usage on protein expression is more significant than the influence of mRNA folding factors.

In addition to the complex influence of ramp sequence during translation initiation, the interplay between codon usage and mRNA structure influences protein expression and host growth by altering the ratio of cellular resources, such as ribosomes and translational essentials. When the 5' end of an mRNA is minimally structured and lacks translation-slowing codons, free ribosomes can rapidly bind and translate, producing high amounts of protein while protecting the mRNA from degradation. The presence of rare codons that slow translation can lead to ribosome stalling, reduced protein production, and increased ribosome occupancy. When only the 5' end of an mRNA is highly structured, ribosomes have difficulty binding and initiating translation, resulting in very low protein expression without significant consumption of nutrients or ribosomes. When both the 5' end and downstream regions are highly structured, ribosomes find it challenging to initiate translation, while the downstream structure protects the mRNA from degradation. This situation consumes a large number of "idle" ribosomes but results in minimal protein production, which is detrimental to both protein expression and host growth. Other factors, such as GC content and potential enzymatic cleavage sites, have also been considered as potential influencing factors.

Solutions and comprehensive optimization methods
To solve the problem of protein non-expression, traditional methods include codon optimization and the use of fusion tags. At the same time, high-throughput experimental technology and artificial intelligence are also booming.
1. Codon Optimization
To overcome challenges in codon usage and mRNA structure that may hinder recombinant protein expression, optimization of the gene sequence is necessary prior to cloning into an E. coli host.
Common codon optimization strategies
1. Using Optimal Codons (UBC): Following the “one amino acid-one codon” approach, the original codons were replaced with the most common host codons or codons with high tAI or nTE values.
2. Matching codon usage (MCU): Adjust the frequency of specific codons in the target gene based on their frequency in the host.
3. Harmonizing relative codon adaptation (HRCA): harmonizing the codon frequency of the original host's genes with the codon frequency of the host.
Codon Optimization Tools
1. OPTIMIZER: It uses a simple "one amino acid-one codon" strategy and combines it with a randomized approach based on the Monte Carlo algorithm to maximize optimization while minimizing sequence changes, thereby improving protein expression levels.
2. DNA Chisel: Allows users to choose among these three optimization strategies, providing researchers with a customized approach to gene sequence optimization.
In addition to the above methods, heterologous tRNAs can be introduced. By introducing plasmids containing tRNA genes corresponding to rare codons, the problem of tRNA depletion that may occur during translation can be solved. For example, the Rosetta strain series carries the pRARE plasmid, which contains tRNA genes that decode rare codons.
Application of fusion tags
In E. coli expression systems, the use of fusion tags/short peptides is a highly effective strategy for enhancing protein expression and addressing the problem of non-expressed proteins. Fusion tags, particularly those located at the N-terminus of proteins, play a crucial role in regulating the nucleotide sequence near the translation initiation region and integrating exogenous functional tags. These fusion tags modify the ramp sequence during translation, enabling high expression of proteins with low expression levels and assisting protein folding. Common fusion tags include MBP, SUMO, and TrxA. On the other hand, smaller fusion peptides, typically consisting of no more than 15 amino acid residues, can also play a significant role in improving expression levels and solubility. Protein solubility is lowest when the environmental pH is equal to the isoelectric point of the protein. Therefore, much research has focused on introducing short peptides composed of charged amino acids to alter the net charge of the target protein to positive or negative, thereby improving its solubility. Selecting an appropriate peptide tag based on the isoelectric point of the target protein can enhance solubility and prevent aggregation.


Comprehensive optimization method based on large-scale data and deep learning
With advances in high-throughput culture technology and deep learning methods, new tools and algorithms have emerged that enable direct codon optimization based on large datasets of highly expressed sequences. These tools leverage large-scale and deep learning techniques to provide more accurate guidance for protein expression. Currently, comprehensive optimization methods used to predict and improve protein expression levels include 6AA/31C, MPEPE, SoluProt, ICOR, COSMO, BiLSTM-CRF, and DeepTESR.

Summary and Outlook
This study illustrates that protein toxicity and gene sequence are two key factors contributing to the failure or low expression of proteins. Toxic protein expression can be controlled by controlling the expression of toxic proteins at different growth stages, selecting appropriate host strains and induction systems, and utilizing secretion expression strategies. The protein gene sequence itself significantly influences its expression, and the mechanisms underlying this influence are complex, with potential influencing factors including codon usage and mRNA structure. The problem of protein non-expression can be addressed through codon optimization and the addition of fusion tags. Meanwhile, advances in artificial intelligence (AI) are enabling researchers to redesign gene sequences from new perspectives. By learning from extensive protein expression databases, AI can identify hidden sequence features and provide sequence predictions that lead to high protein expression. Leveraging AI remains a viable approach, and the latest large-scale language models can already facilitate de novo protein design. It is anticipated that in the future, nucleic acid-based design of target protein gene sequences will also become possible.

