Background
G protein-coupled receptors (GPCRs) are important therapeutic targets, but their complex structures pose challenges for effective drug design. Nanobodies (i.e., single-domain antibodies) have emerged as a promising therapeutic strategy for targeting GPCRs, offering advantages over traditional small molecules and antibodies. However, an incomplete understanding of the structural features that enable GPCR-nanobody interactions has limited their development.
On May 30, 2024, American researchers published an article titled " Structural basis for selectivity and antagonism in extracellular GPCR-nanobodies " in Nature communications. The researchers studied VUN701, a nanobody antagonist targeting atypical chemokine receptor 3 (ACKR3). They determined that ACKR3 binding requires an extended CDR3 loop. CDR3 is uncommon in most nanobodies, but is prevalent in nanobodies targeting GPCRs. Combining experimental and computational methods, the researchers mapped an inhibitory ACKR3-VUN701 interface and defined a unique conformational mechanism for GPCR inactivation. The results provide insights into the selectivity of class A GPCR-nanobodies and provide strategies for the development of these new therapeutic tools.

Development of G protein-coupled receptor (GPCR) targeted therapies
Over 800 human G protein-coupled receptors (GPCRs) allow cells throughout the body to respond to countless extracellular signals. The exquisite regulation of these receptors makes them one of the most abundant classes of therapeutic targets. However, only one in eight GPCRs has been successfully targeted by current therapeutic strategies. Monoclonal antibodies (mAbs) have been explored as a means of overcoming the challenges of GPCR therapeutic targeting. Two mAbs targeting GPCRs have been approved by the FDA (erenumab for CGRP-R and mogamulizumab for CCR4). While monoclonal antibody targeting has achieved some success, the expanded binding surface of mAbs is not always well suited for selective recognition of the relatively small extracellular epitopes displayed by membrane-embedded GPCRs. Nanobodies (Nanobodies) have the potential to bridge the gap between monoclonal antibodies and small molecules. The approval of the first nanobody (placizumab) in 2019 has accelerated the development of this new class of therapeutic molecules. These small (12-15 kDa), easily produced proteins bind to GPCR epitopes that are more difficult to target with monoclonal antibodies using a smaller counterpart composed of three complementarity-determining regions (CDRs). Nanobodies targeting two GPCRs (chemokine receptors CXCR4 and CX3CR1) are currently in clinical trials. To fully exploit the potential of nanobodies as pharmacological tools and potential drugs, it is crucial to understand how they bind to extracellular GPCR epitopes and alter receptor signaling. We will focus on the most abundant GPCR targets, class A GPCRs. Of the over 1,900 solved nanobody structures, only 2% target class A GPCRs, the majority of which bind to the intracellular surface of the receptors. Of these five anti-GPCR nanobodies that bind to the extracellular surface, only two are pharmacologically active. These nanobodies, JN241 and NbE, are neutral antagonists of the apelin receptor and the μ-opioid receptor, respectively. JN241 has also been designed as an apelin receptor agonist (JN241-Y). Several other nanobodies have been identified as extracellular antagonists, but details of the nanobody-GPCR interface and the structural basis of receptor inhibition are lacking.

VUN701 selectivity and its mechanism
Using a BRET assay to measure β-arrestin2 recruitment to ACKR3, we found that CXCL11-dependent signaling was inhibited by VUN701; VUN701 also inhibited CXCL12 activation.

Schild analysis of CXCL12-induced β-arrestin2 recruitment to ACKR3 demonstrated that VUN701 altered CXCL12 potency in a concentration-dependent manner without altering the slope or maximal potency of CXCL12 β-arrestin2 recruitment. CXCR4 and CXCR3 are receptors for CXCL12 and CXCL11, respectively. In summary, the inhibitory effects of VUN701 on CXCL11 and CXCL12 function in cell-based assays, as well as Schild analysis of CXCL12, suggest that VUN701 acts as a selective, reversible, competitive antagonist. Direct competition by VUN701 with both CXCL11 and CXCL12 suggests that these two ligands share overlapping orthosteric epitopes.

CDR3 structural features of VUN701:
The researchers analyzed the solution structure of VUN701 by NMR. VUN701 adopts a conserved β-sandwich immunoglobulin fold observed in other nanobody structures, with three variable loops, including the complementarity determining regions (CDRs), which mainly mediate binding to the target protein. The extended β-hairpin conformation observed for CDR3 was confirmed by the presence of strong cross-chain nuclear Overhauser effect (NOE) signals, low root mean square deviation (RMSD) values, and high heteronuclear NOE ratios for residues 91–101 and 105–109.
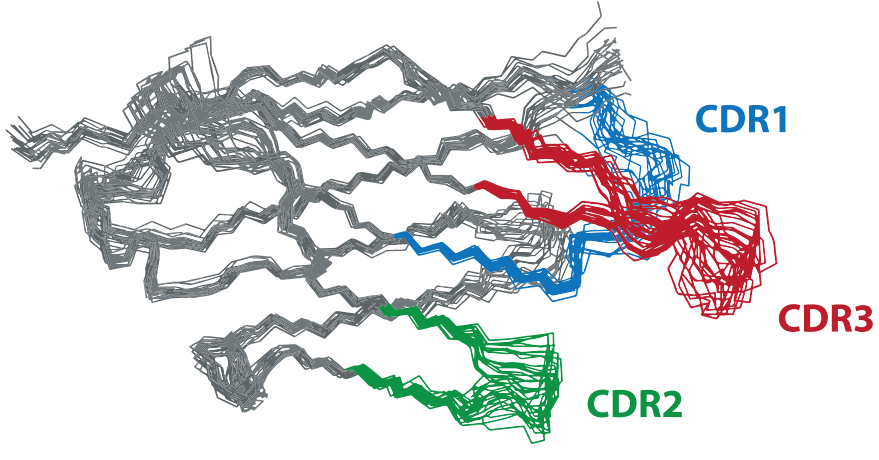
The extended CDR3 of VUN701 is a unique feature in the solved nanobody structures. The 1962 solved nanobody structures have three types of CD3: folded (66%), extended (25%) or short (9%).

Different CDR3 distributions were observed for class A GPCR-targeting nanobodies, of which nearly 70% belonged to the extended class. We speculated that the extended CDR3 " β finger " of VUN701 was directly involved in the binding of high-affinity ACKR3, and replaced each CDR of VUN701 to evaluate their respective contributions.


While the ΔCDR1 and ΔCDR2 mutants retained the ability to bind and inhibit ACKR3, the ΔCDR3 mutant resulted in a near-complete loss of inhibition. The functional importance of the β-finger and the enrichment of extended CDR3s in nanobodies that bind to GPCRs led the researchers to hypothesize that VUN701 binds to the orthosteric pocket of ACKR3 and interacts in a manner that mimics the N-terminal interactions of chemokine ligands.

Mapping of the VUN701-ACKR3 interface
Using AlphaFold2 and molecular dynamics simulations, the researchers constructed models of ACKR3 in complex with its natural ligand CXCL12 and VUN701. The CDR3 loop of VUN701 inserts into the orthogonal site of ACKR3 in an extended β-hairpin conformation. This insertion is a key feature of VUN701 binding to ACKR3 and is relatively common in GPCR-targeting nanobodies.

The researchers used modeling to predict potential contact points between VUN701 and ACKR3 and experimentally verified the importance of these contact points. In particular, they found that certain amino acid residues in the CDR3 loop (such as K100 and R103) formed key interactions with specific residues in ACKR3 (such as D2756x58 and E2907x27).

To assess the importance of ACKR3 contacts, the researchers selected 14 charged aromatic residues lining the chemokine binding pocket for alanine substitutions. The researchers found functional effects of the ACKR3 substitutions: five of the substitutions had little effect on CXCL12 potency, while the other six substitutions altered the EC50 by 2- to 100-fold.

ACKR3 antagonistic mechanism
Structural comparisons of ACKR3 bound to the chemokine agonist CXCL12 and the nanobody antagonist VUN701 reveal key contacts or conformational changes that underlie receptor inactivation. The most significant difference between the VUN701-ACKR3 model and the CXCL12-ACKR3 cryo-electron microscopy structure (PDB: 7SK5) is a significant displacement of transmembrane helix 7 (TM7). VUN701 binding appears to position CDR1 and CDR2 directly above TM7, forming a lid that prevents TM7 from occupying the position observed in the CXCL12-ACKR3 structure. The CXCL12-induced upward shift of TM7 is associated with a conserved network of polar side chains that stabilizes the active conformation. This network includes the NPxxY motif in TM7 that interacts with D902x50. VUN701 binding prevents the upward shift of TM7, which is required for binding to the conserved NPxxY activation motif.

VUN701 selectivity
Using BRET assays, the researchers measured the functional selectivity of VUN701 for the ACKR3 receptor. The results showed that VUN701 effectively inhibited ACKR3 binding to CXCL11 and CXCL12 with nanomolar IC50 values, but had no effect on β-arrestin2 recruitment to CXCR4 or CXCR3, demonstrating VUN701's high selectivity for ACKR3. Comparing sequence conservation across different chemokine receptors revealed that residues within the transmembrane helices and orthogonal sites were more conserved, while residues in the N-terminus, ECL2, and ECL3 regions were more diverse. This suggests that VUN701's selectivity may be related to its ability to bind to residues in a specific region of ACKR3. The CDR1 and CDR2 loops of VUN701 interact with specific regions of ACKR3, contributing significantly to VUN701's selectivity. The selectivity of VUN701 is determined not only by the contact points within the orthogonal site but also by its long-range contacts with the unique region of ACKR3, which provides a structural basis for the design of new GPCR-targeting nanobodies.

Summary and Outlook
This study deeply analyzed the interaction of the VUN701 nanobody with atypical chemokine receptor 3 (ACKR3), revealing the structural basis for its high selectivity and antagonistic effects. VUN701 utilizes its unique extended CDR3 β-hairpin structure to bind to an orthogonal site on ACKR3, effectively blocking receptor activation and demonstrating high selectivity. These findings provide new insights into GPCR-nanobody interactions and a structural basis for the development of novel GPCR-targeted drugs, particularly nanobodies that act as agonists or antagonists against specific GPCRs.
