Background
The correct assembly of a four-chain IgG-like bispecific composed of two different light and heavy chains requires the formation of three heterodimer interfaces: one formed by the two different heavy chain (HC) Fc domains and two Fab heterodimers formed by homologous pairing of heavy and light chains (LC). Without a strong driving force, random interchain interactions lead to the formation of heavy chain homodimers and mispairing between the light and heavy chains. Correct pairing of cognate heavy and light chains is crucial for the efficient production of IgG-like bispecific antibodies (bsAbs) from a single host cell.

On March 24, 2025, Adimab published an article titled "Design of orthogonal constant domain interfaces to aid proper heavy/light chain pairing of bispecific antibodies" in MAbs. The article proposes a universal solution for eliminating heavy chain (HC):light chain (LC) mispairing in bispecific antibodies by using two orthogonal constant domain (CH1:Cκ) interfaces containing computationally designed amino acid substitutions. Rosetta designed a new hydrogen bond (H-bond) network at the CH1:Cκ interface, and then Rosetta energy calculations identified designs with enhanced pairing specificity and interface stability. The final design, consisting of 11 amino acid substitutions in both Fab constant regions, was tested on six IgG-like bsAbs with various unmodified human antibody variable domains. These engineered bispecific IgG molecules also exhibited high expression titers in mammalian cell lines, good developmental properties, and no increased immunogenicity as assessed by in vitro immunogenicity assays. This computationally designed constant domain mutation set provides a broadly applicable approach for bispecific antibody production, potentially improving the efficiency and reliability of therapeutic antibody production.

Single Interface Design (SID)
A single-interface design (SID) four-chain bsAb consists of a wild-type CH1:Ck domain interface on one Fab arm and a mutant CH1:Cκ domain interface on the other. The researchers used Rosetta's HBNet algorithm to design 3,571 CH1:Cκ mutation combinations and identified 172 of 1469 designs with high pairing specificity using flex ddG energy calculations.

Using HEK293 cells for transient expression, 18 of the 20 designs were successfully expressed and analyzed by LC-MS. The results showed that eight designs achieved correct pairing rates exceeding 79% compared to the wild-type (WT) design (without constant domain mutations). Designs 1039 and 1443 achieved correct pairing rates of 88% and 86%, respectively.

Dual Interface Design (DID)
Dual-Interface Design (DID) bsAbs contain mutations in the CH1:Cκ domain interface on both Fab arms. Two designs (designs 1443 and 1039) that were typically among the best-scoring design combinations were selected as design "A" (one of the mutation sets of the two CH1:Cκ designs present in DID) in the context of the ustekinumab variable domain sequence. These two designs were screened against five designs (design "B") that were cloned in the context of the ustekinumab variable domain sequence and further screened using Rosetta energy calculations.
Ten DID designs were expressed in HEK-293 cells, and the results showed that all designs had a higher correct pairing rate than the wild-type (71%). Specifically, the correct pairing rate for the design 1443/1993 mutation reached 100% (verified by LC-MS), and the main peak ratio in CEX analysis was as high as 88%.

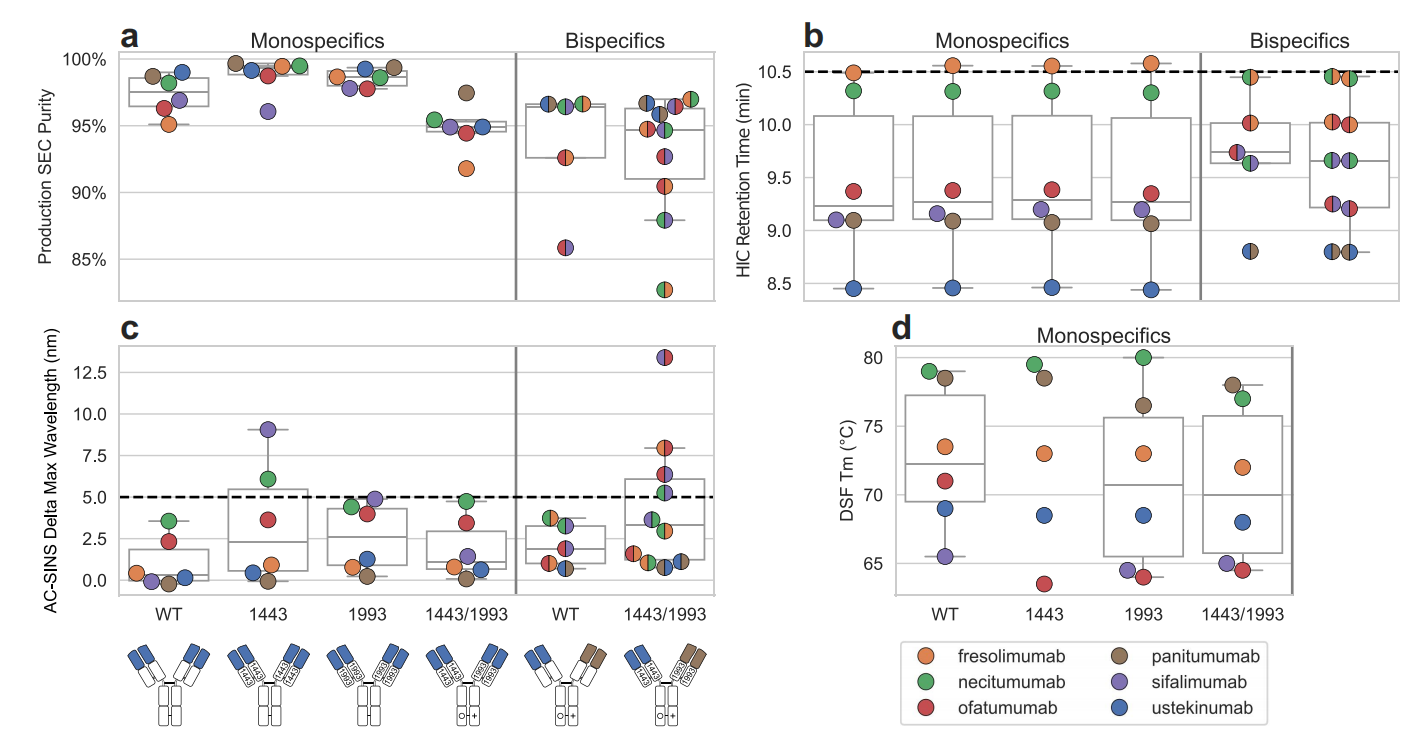
Developability Assessment of CHO-Produced Bispecific Antibodies
Five bsAbs were produced using a Chinese hamster ovary (CHO) cell line, and purity was further assessed by SEC. Monospecific controls with either a WT constant domain or a Fab arm containing only one of the 1443 or 1993 mutation sets (and a CH3 KiH) typically returned SEC purities of >97%.
The HIC retention time was not affected by the introduction of either the 1443 or 1993 pairwise mutations, and the HIC value of the bispecific antibody was observed to correspond to the average of its two constituent monospecific values, regardless of whether CH1:Cκ was mutated.
AC-SINS analysis showed that the mutational design slightly increased self-interactions, but did not exceed the threshold for clinical advancement (5 nM). This suggests that the mutational design did not significantly affect the antibody's self-interaction properties. As with SEC purity, the directionality of the variable and constant region mutation sets was also important, with bispecific molecules displaying different AC-SINS values depending on their orientation.
The stability of the Fab fragments was assessed by differential scanning fluorimetry (DSF), and the mutational design had minimal impact on the thermal stability and aggregation temperature of the Fab. Although the Fab Tm and Tagg of some combinations decreased slightly, these changes did not exceed acceptable limits.
After the introduction of design sets 1443 or 1993, the total yield of bispecific samples produced by transient expression in CHO cells decreased from 212 mg/L for bispecific samples produced using the WT CH1:Cκ constant region to 185 mg/L for samples produced using DID 1443/1993.

In vitro assessment of immunogenicity risk
In vitro evaluation of monoclonal antibodies carrying the 1443/1993 constant domain mutation set found no evidence that these mutations confer an additional immunogenicity risk.

Crystal structure and structural analysis
The structures of Fabs containing designed mutations 1443 and 1993 were solved using X-ray crystallography. The crystal structures revealed that the designed mutations introduced a new hydrogen-bonding network, with the mutated residues primarily located in the core region of the CH1:Cκ interface. The RMSDs of the structures of designs 1443 and 1993 compared to the wild-type were 0.45 Å and 0.43 Å, respectively, indicating that the mutations did not induce significant structural rearrangements.

Summarize
This article presents a set of computationally designed constant domain mutations that enforce correct heavy/light chain pairing in multiple tested variable domain contexts in a single-cell host, suggesting the value of producing correctly assembled bispecific antibodies in downstream manufacturing and production settings. High-throughput screening assays did not reveal a general increase in downstream developmental risk, nor did in vitro immunogenicity assays. Crystal structures of the 1443 and 1993 mutation sets revealed the presence of several new hydrogen bonds introduced by these designs. The computationally designed hydrogen bond network significantly enhanced the specificity and stability of the protein interface. This design approach, including hydrogen bond network design and polymorphic rescoring, may serve as a general template for designing other protein-protein interfaces where preferential pairing is desired.
