Background
In addition to their widespread application in biological research, such as protein purification and imaging, recombinant fusion proteins have also become a significant category of biopharmaceuticals. For example, many protein therapeutics are fused to the Fc region of antibodies or to carrier proteins to extend their plasma half-life and enhance their therapeutic efficacy. Fusion proteins can also be used for drug targeting and delivery. With the rapid advancement of biotechnology, fusion protein technology is becoming increasingly important in creating novel protein therapeutics and improving the performance of existing protein drugs. The successful construction of recombinant fusion proteins requires two essential elements: the fusion protein and the linker. Selecting an appropriate linker to connect protein domains can be complex and is often overlooked in fusion protein design. Direct fusion of functional domains without a linker can lead to numerous undesirable outcomes, including misfolding of the fusion protein, low protein yield, or impaired biological activity. Therefore, the selection or rational design of linkers to connect fusion protein domains is an important yet underexplored area of recombinant fusion protein technology.

An article titled "Fusion protein linkers: property, design, and functionality" published in Adv Drug Deliv Rev provides an overview of the properties of linkers in naturally occurring multi-domain proteins and discusses, with examples, empirical linkers that have been successfully applied to the construction of recombinant fusion proteins. Finally, it describes the various functions that can be achieved by utilizing linkers in recombinant fusion proteins, including improved folding and stability, enhanced protein expression, increased intrinsic bioactivity, the ability to target specific sites in the body, and the ability to alter the pharmacokinetic (PK) profile of fusion proteins.

General properties of linkers in natural multidomain proteins
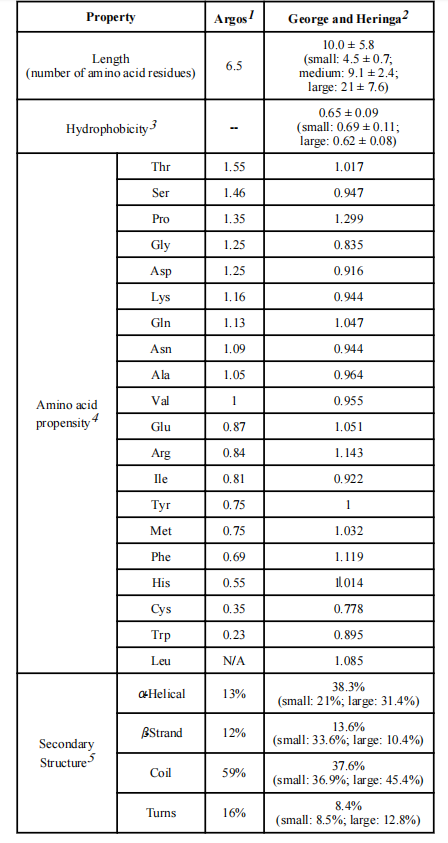
Naturally occurring multidomain proteins consist of two or more functional domains connected by a linker peptide. These linkers serve to connect protein segments and also provide numerous other functions, such as maintaining cooperative interdomain interactions or preserving biological activity. Studies have found that increasing linker length leads to greater solvent accessibility. The average hydrophobicity of the linker decreases with increasing length, indicating that longer linkers are more hydrophilic.
Amino acid residue preferences in natural linkers: Polar uncharged or charged residues are preferred, with proline (Pro), threonine (Thr), and glutamine (Gln) being ideal amino acids for natural linkers. Proline is a unique amino acid with a cyclic side chain, resulting in highly constrained conformations. The lack of amide hydrogen in proline may prevent hydrogen bonding with other amino acids, thereby reducing interactions between the linker and the protein domain. Therefore, the inclusion of a proline residue may increase the rigidity and structural independence of the linker.
Natural linkers adopt various conformations in their secondary structure: Such as helices, β-strands, coils/bends, and turns, to perform their functions. Based on the secondary structure analysis of George and Heringa, linkers are divided into two categories, helical and non-helical. Linkers with α-helical structures may effectively separate protein domains by acting as rigid spacers and reducing their adverse interactions. Consequently, this conformation is commonly adopted by many natural and empirical linkers . Non-helical linkers, which are rich in Pro sequences, can exhibit relatively rigid structures and help reduce interdomain interference.
Linker in recombinant fusion protein
Linkers in recombinant fusion proteins are mainly divided into flexible linkers, rigid linkers and cleavable linkers, which play different functions in the construction of fusion proteins.
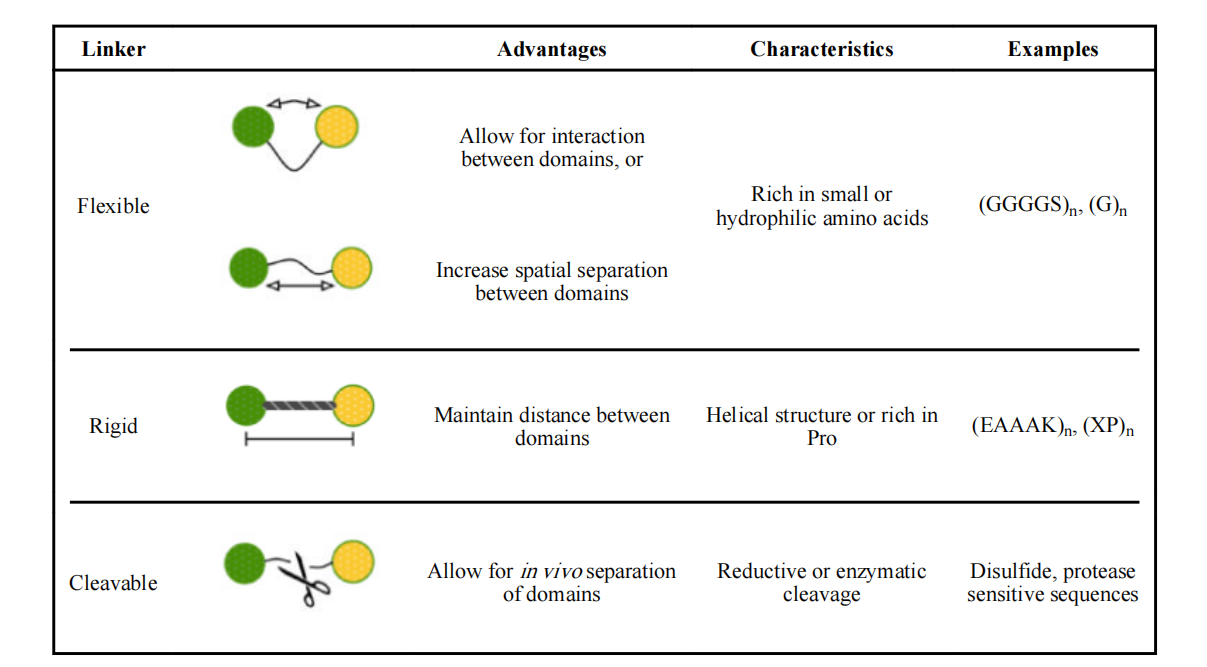
Flexible linker
Flexible linkers are typically composed of small, nonpolar (glycine) or polar (serine or threonine) amino acids. The small size of these amino acids provides flexibility, enabling mobility of the linked functional domains. The addition of serine or threonine maintains the stability of the linker in aqueous solution, thereby reducing adverse interactions between the linker and the protein moiety. Polar amino acids, such as lysine and glutamate, are added to enhance solubility.
Commonly used flexible linkers: For example (GGGGS)n; (G)n; KESGSVSSEQLAQFRSLD and EGKSSGSGSESKST;GSAGSAAGSGEF.
Advantages of flexible linker: They can be used to maintain the distance between functional domains, and the length of flexible linkers can be adjusted to facilitate proper folding or achieve optimal biological activity of the fusion protein.
Disadvantages of flexible linkers: Linkers lack rigidity. Because flexible linkers are relatively short and flexible, undesirable interactions or interferences may occur between the connected protein domains, which may lead to low expression yields or loss of biological activity.
Rigid linker
Rigid linkers typically adopt an α-helical structure or contain multiple Pro residues, exhibiting a relatively rigid structure. In many cases, they are more effective at separating functional domains than flexible linkers. Linker length can be easily adjusted by varying the copy number to achieve the optimal distance between domains. Rigid linkers are selected when spatial separation of domains is critical for maintaining the stability and biological activity of the fusion protein.
Commonly used rigid linker (EAAAK)n sequences; A(EAAAK)n A (n=2-5) is an α-helical conformation; proline-rich sequence (XP)n, where X represents any amino acid, preferably alanine, lysine or glutamate. The presence of proline in non-helical linkers can increase rigidity and allow effective separation of protein domains.
In vivo cleavable linker
In vivo cleavable linkers utilize unique in vivo processes and are cleaved under specific conditions, such as in the presence of reducing agents or proteases. This type of linker may reduce steric hindrance, enhance biological activity, or enable independent function/metabolism of the functional domains of the recombinant fusion protein after linker cleavage.
Reduction of disulfide bonds: The disulfide linker (LEAGCKNFFPR↓SFTSCGSLE) designed by taking advantage of the reversibility of disulfide bonds is based on a disulfide cyclic peptide containing an internal disulfide bond formed between two cysteine (Cys) residues and a thrombin-sensitive sequence (PRS) between the two Cys residues.
Specific protease cleavage: In vivo cleavage of the linker in a recombinant fusion protein can also be achieved by proteases expressed in specific cells or tissues or in certain cellular compartments under pathological conditions (e.g., cancer or inflammation), thereby releasing the free active domain in vivo.

Linker in fusion protein
The most fundamental function of a linker in a recombinant fusion protein is to covalently connect functional domains (flexible or rigid linkers) or release them under desired conditions (cleavable linkers). Linkers can also provide numerous derivative functions in protein drug design, such as enhancing biological activity, increasing production, enabling controlled or targeted drug delivery, and achieving desirable pharmacokinetic properties for the fusion protein.

Linker improves folding and stability of fusion proteins
Flexible GS linker: It can be used to construct single-chain variable fragments (scFv). Its high flexibility allows the VH and VL domains to be correctly positioned without interfering with protein domain folding. By adjusting the length of the GS linker , the distance between the VH and VL domains can be optimized, thereby achieving correct folding of the functional domains and the necessary interactions.
(G)n linker: Insertion of a ( Gly)8 linker between the Myc epitope tag and the protein of interest (Est2p) significantly improved the functionality of the epitope-tagged Est2p. This improvement was attributed to proper folding after linker insertion and reduced steric hindrance between the functional domains.
Alpha-helical linkers: Such as (EAAAK)n, provide sufficient distance to allow independent folding of protein domains and facilitate the correct assembly of viral particles.
Linker length and structure on stability: As flexible linker length increased, the stability of the fusion protein improved. α-helical linkers appeared to be more effective in improving thermal stability compared to flexible linkers , likely because the rigid structure of α-helical linkers provides ample space for protein domains to fold and function independently.
Linker improves fusion protein expression
Linkers can reduce interference between different functional domains in a fusion protein, promote the independent folding of each domain, and thus improve the overall expression level of the fusion protein. By providing appropriate space and structure, linkers enable the fusion protein to fold correctly and stabilize in the endoplasmic reticulum, reducing misfolding and instability caused by domain interference, thereby improving expression.
Linkers can enhance the biological activity of fusion proteins
In fusion proteins, linkers can provide an appropriate distance between domains to reduce their interference, restore or improve folding, or allow for the in vivo release of free protein drug domains, ultimately enhancing bioactivity. For example, the insertion of an α-helical linker ( e.g., A(EAAAK)4ALEA(EAAAK)4A) in a fusion protein of granulocyte colony-stimulating factor (G-CSF) and transferrin (Tf) significantly enhanced the in vitro bioactivity of G-CSF and demonstrated enhanced in vivo efficacy in animal models via oral administration.

Linkers can target fusion proteins to specific sites in vivo
Linkers can improve the targeting of fusion proteins by increasing the binding affinity of the targeted protein domain to its receptor or by releasing the active protein domain through specific cleavage at a specific site.
Targeting under physiological or pathological conditions: In vivo cleavable linkers that are sensitive to proteases that become active under certain physiological or pathological conditions are ideal for drug targeting. For example, coagulation factor IX (FIX) is fused to albumin, and the linker is specifically cleaved by proteases that are activated during coagulation , thereby releasing active FIX at the coagulation site.
Disease-Specific Targeting for Overexpressed Proteins: The linker can contain a cleavage site for a protease overexpressed at the disease site, allowing the fusion protein to be specifically cleaved at the disease site, releasing the active protein domain. For example, matrix metalloproteinases (MMPs) are overexpressed in various diseases. By inserting an MMP-sensitive linker into the fusion protein, specific activation at the disease site can be achieved.
Disease-specific targeting with specific expression: The linker can be designed to contain a cleavage site for a protease expressed by a specific pathogen, allowing the fusion protein to be activated upon encountering that specific pathogen.
Intracellular targeting: In addition to the extracellular space, the targeting/activation site of the in vivo cleavable linker can also be located inside the cell. For example, by utilizing proteases active in the endoplasmic reticulum or lysosomes, such as furin or cathepsin B, the fusion protein can be cleaved at a specific location after entering the cell, releasing the active protein domain.

Linker affects PK of fusion protein
Bifunctional fusion proteins have a much more complex PK/PD profile because their distribution is influenced by two distinct domains/binding sites. Linker insertion may alter the receptor binding affinity of each protein domain, potentially affecting the in vivo disposition of the fusion protein and increasing the complexity of PK studies.
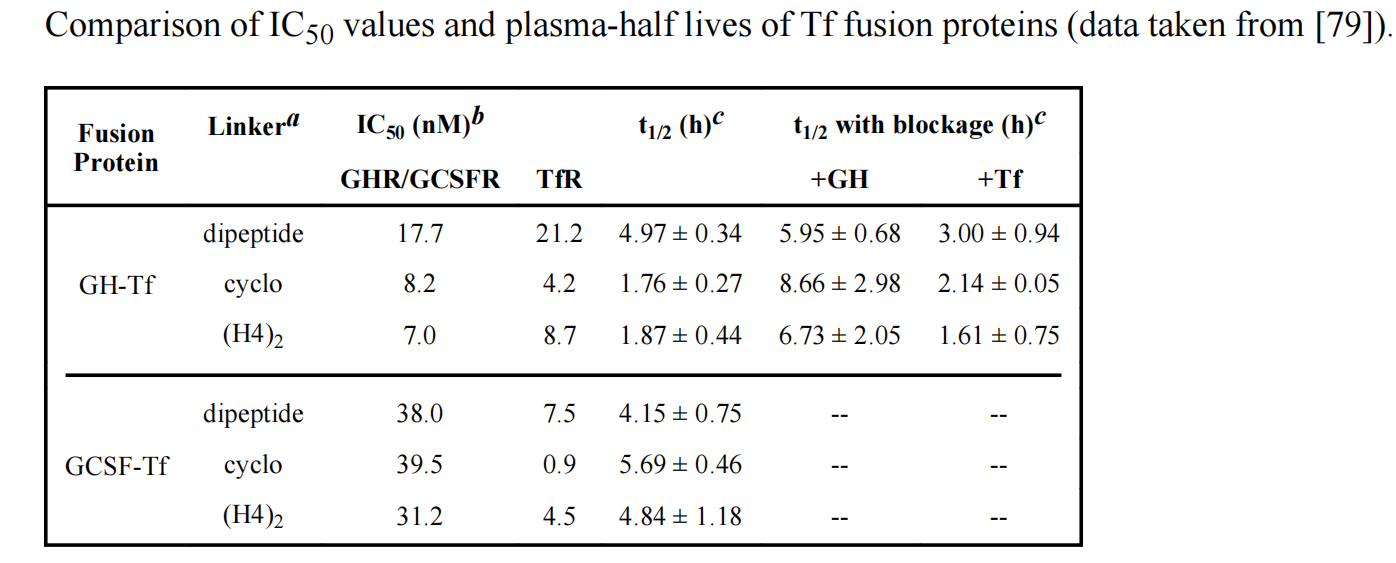
The relative binding affinity of each protein domain to its receptor (which can vary with different linkers ) can affect the half-life of the fusion protein.

Summarize
Linkers primarily include flexible linkers, rigid linkers, and in vivo cleavable linkers. Optimal linkers can offer numerous advantages for fusion protein production, including improved structural stability, enhanced bioactivity, increased expression levels, altered PK profiles, and in vivo targeting of fusion proteins. With the rapid advancements in protein science and biotechnology, linker design in fusion proteins has become more important than ever. As future biomedical research deepens its understanding of structure, conformation, and function, the incorporation of linkers will greatly facilitate the construction of stable and bioactive recombinant fusion proteins for drug delivery applications.
