Protein-protein interactions are fundamental to cellular structure and function. Most proteins do not function in isolation but rather through interactions with other proteins. These interactions mediate biological processes such as catalysis, transport, and signal transduction, playing a crucial role in understanding protein function, disease pathogenesis, and drug development. Commonly used protein-protein interaction methods include surface plasmon resonance (SPR), co-immunoprecipitation (Co-IP), GST pull-down, immunofluorescence (IF), colocalized yeast two-hybrid (Y2H) system, fluorescence resonance energy transfer (FRET), proximity labeling (PL), bimolecular fluorescence complementation (BiFC), isothermal titration calorimetry (ITC), and double electron-electron resonance spectroscopy (DEER). This article provides a brief overview and comparison of these methods.
Surface plasmon resonance (SPR)
SPR (surface plasmon resonance) is an analytical technique used to study protein-protein interactions. Its basic principle is to exploit the phenomenon of total internal reflection of light on the surfaces of prisms and metal films. When a beam of light at a specific angle strikes a thin metal film (such as a gold film), the free electrons in the metal interact with the light wave, forming a phenomenon known as surface plasmon resonance (SPR). This resonance causes a significant decrease in the intensity of the reflected light at a specific angle (i.e., the SPR angle). Because the SPR state is extremely sensitive to changes in the refractive index of the medium near the metal surface, when biomolecules bind to the metal surface, this causes a change in the local refractive index, which in turn causes a shift in the SPR angle. By precisely measuring this angle change, we can monitor interactions between biomolecules in real time, including the binding and dissociation processes.
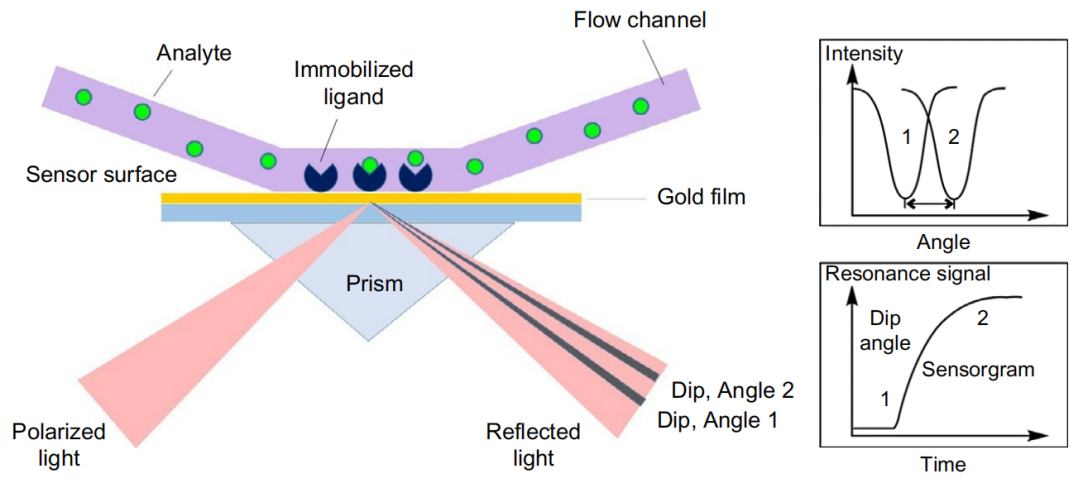
(Data source: Drescher DG, et al. Adv Protein Chem Struct Biol. 2018)
Co-immunoprecipitation (co-IP)
Co-immunoprecipitation (Co-IP) is widely used to identify and validate protein-protein interactions (PPIs). Based on the specific immunological interaction between a bait protein and its antibody, Co-IP has become a reliable and effective method for detecting physiological interactions between proteins.
When cells are lysed under non-denaturing conditions, many intracellular protein-protein interactions are preserved. For example, a bait protein (e.g., protein X) can be captured by stabilizing it on agarose beads using a specific antibody. If another protein, a prey protein (e.g., protein Y), binds to protein X in vivo, the antibody can be used to co-precipitate the protein X-protein Y complex. Subsequently, by investigating protein Y, we can confirm protein X-protein Y interactions or discover new interactors of protein X.
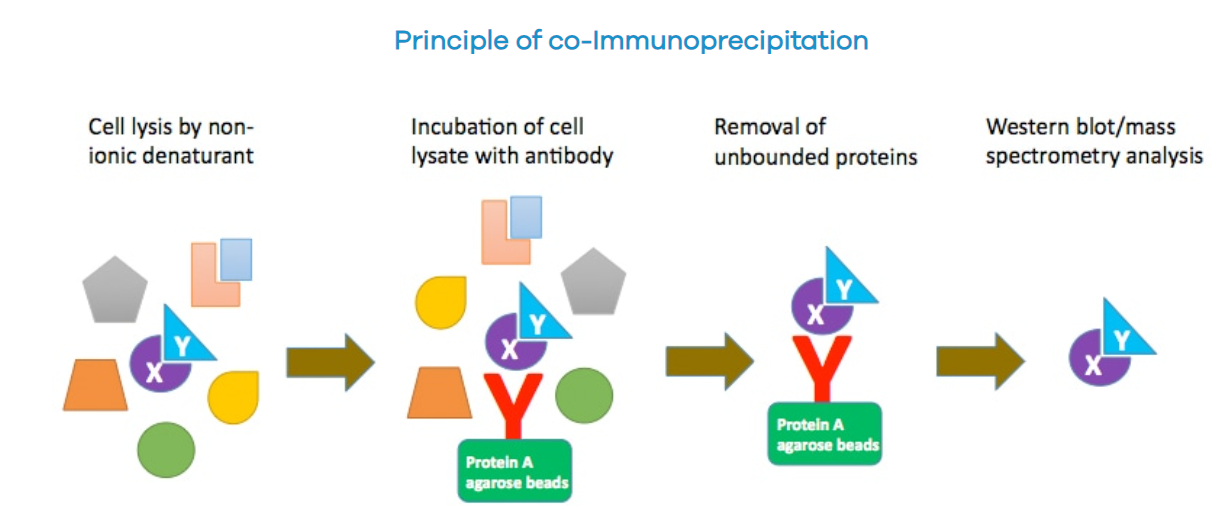
(Data source: profacgen official website)
GST-pull down
The GST-pull down assay is an in vitro affinity purification method that uses a bait protein to enrich proteins that interact with the bait protein.
It utilizes a fusion protein tag (such as a GST tag, His tag, and biotin tag) immobilized on an affinity resin as a bait protein. When the target protein or cell lysate flows through, proteins bound to the bait protein (prey protein) can be captured and "pulled down." Subsequent elution and analysis using Western blot or mass spectrometry (MS) can be used to discover existing protein-protein interactions or conduct preliminary screening to identify new protein-protein interactions.
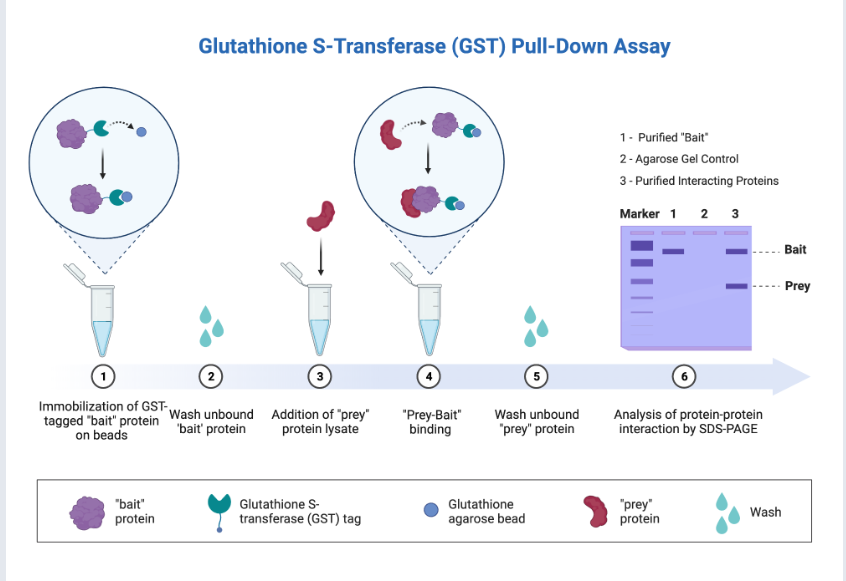
(Data source: BioRender)
Immunofluorescence (IF) colocalization
Immunofluorescence (IF) colocalization combines immunological methods (antigen-antibody specific binding) with fluorescent labeling to study the intracellular distribution of specific protein antigens. It is generally used to verify the colocalization of two or three proteins. Because the fluorescence emitted by fluorescein can be detected under a fluorescence microscope, the cellular localization of antigens can be determined. Two different proteins are labeled with different colored fluorescent markers with different properties. Colocalization is determined by observing the color changes, which can also be analyzed using software.

(Data source Ruiz-Gómez G, Vogel S, Möller S, Pisabarro MT, Hempel U. Sci Rep. 2019)
Yeast two-hybrid system (Y2H)
The yeast two-hybrid system utilizes two independent domains of eukaryotic transcriptional activators: the DNA-binding domain (BD) and the transcriptional activation domain (AD). Transcription is only possible when these two domains function together. In the yeast two-hybrid system, a "bait" protein and a "prey" protein are fused to the AD and BD, respectively, to form two fusion proteins. If these two proteins interact in yeast cells, they bring the AD and BD into close proximity, recreating a functional transcriptional activator and activating expression of a reporter gene.
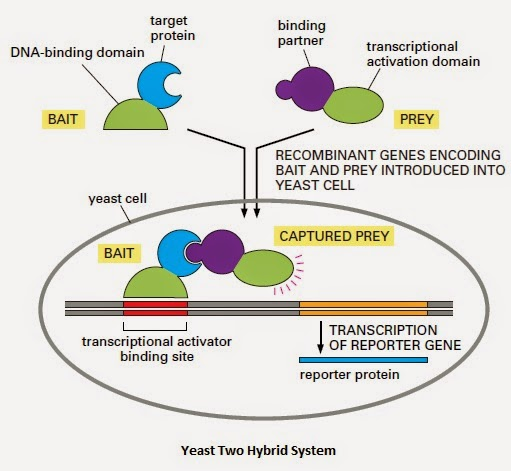
(The pictures used in the data source are from the Internet. If there is any infringement, please contact us to delete them.)
Fluorescence resonance energy transfer ( FRET )
Fluorescence resonance energy transfer (FRET) is an early-developed technology. With the development of green fluorescent protein (GFP) technology, FRET has become a powerful tool for detecting nanoscale distance changes in biomacromolecules, with widespread application in in vivo immunoassays. FRET is the energy transfer phenomenon between two fluorescent molecules in close proximity: the donor and the acceptor . When the emission spectrum of the donor molecule overlaps with the absorption spectrum of the acceptor molecule, and the distance between the two molecules is within 10 nm, non-radioactive energy transfer, or FRET, occurs. This resonant energy transfer to the acceptor causes the donor's fluorescence intensity to be much lower than when it is alone (fluorescence quenching), while the fluorescence emitted by the acceptor is greatly enhanced (sensitized fluorescence). FRET can be performed in living cells without the need for labeling, and it exhibits high sensitivity and specificity.
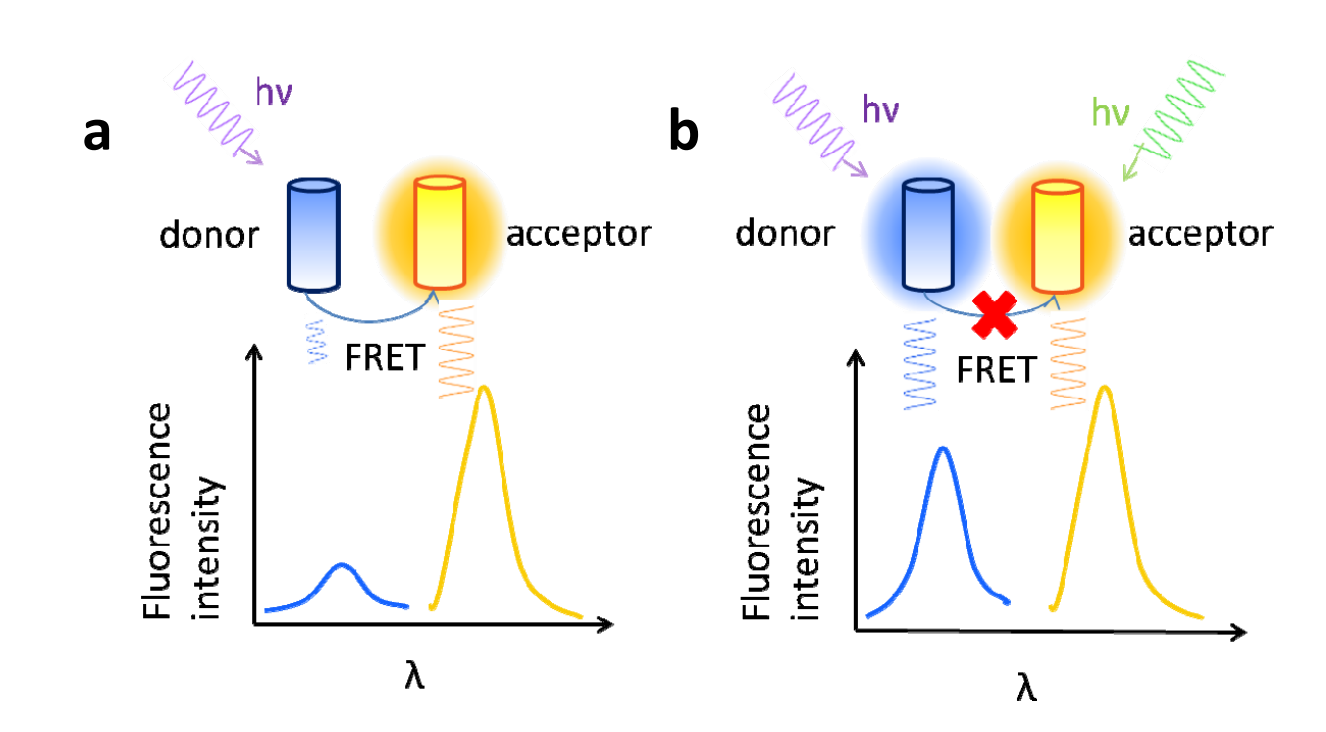
(Data source: Xu F, Wei L, Chen Z, Min W. Opt Express. 2013)
Bimolecular fluorescence complementation (BiFC)
Bimolecular fluorescence complementation (BiFC) analysis enables direct visualization of protein interactions in living cells. BiFC is based on the cleavage and recombination of fluorescent proteins, which are typically divided into N-terminal and C-terminal fragments. When two target proteins interact, the N- and C-terminal fragments of the fluorescent proteins approach each other and recombine to restore their original structure and properties, generating a fluorescent signal that reflects the target protein interaction and provides substantial assistance for examining cell signaling pathways . BiFC has made significant contributions to the study of cell signaling pathways by enabling the examination of interactions between signal transduction proteins and their downstream effector proteins, as well as the analysis of intracellular signaling.
In addition, many other fluorescence complementation technologies have been derived based on BiFC technology, such as mcBiFC, TriFC, dcas13a-SunTag-BiFC, and BiFC-TALE.
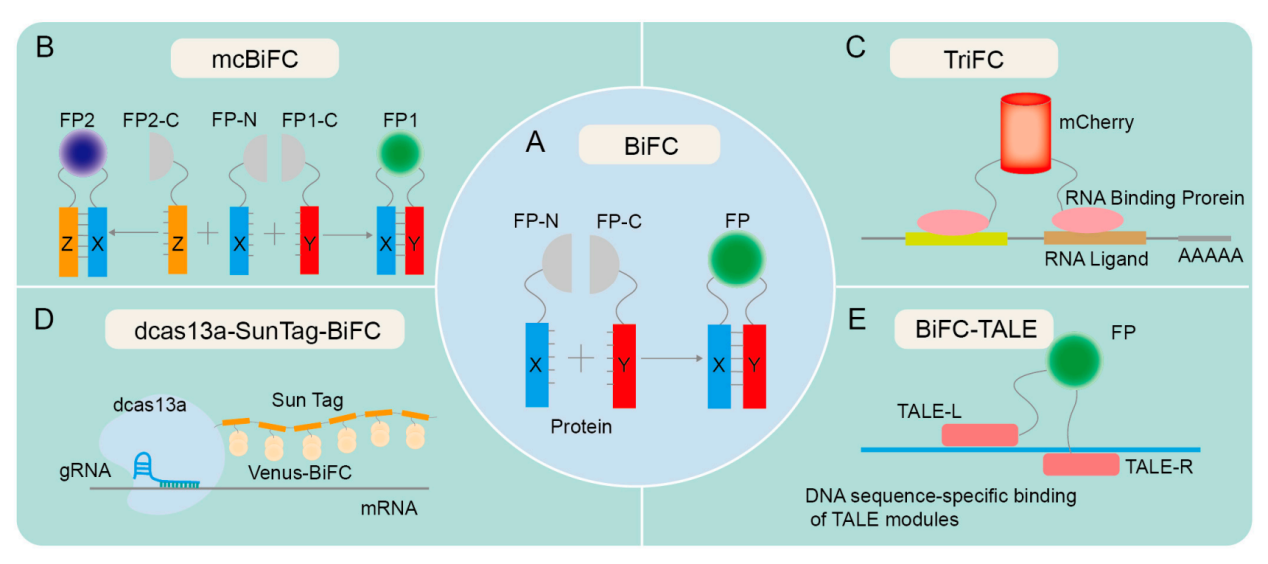
(Data source: Ren H, et al. Biomolecules. 2024)
Isothermal titration calorimetry (ITC)
Isothermal titration calorimetry (ITC) is a technique used to quantitatively study a variety of molecular interactions. This method has been widely applied to investigate the interactions of macromolecules (typically proteins) with small ligands, other proteins, nucleic acids, and drugs. It works by directly measuring the heat released or absorbed during a molecular binding event. In an ITC experiment, a reactant is placed in a temperature-controlled sample cell, connected to a reference cell via a heat flow loop. Both cells are exposed to the same external environment. A specific titrant (e.g., a naturally occurring active compound) is gradually injected into the sample cell. As the sample and titrant react, heat is absorbed or released. With each injection, the heat absorbed or released results in a temperature change between the sample and reference cells. Software integrated with the isothermal titration calorimeter allows data fitting to determine parameters such as binding constants, stoichiometry (n), and enthalpy changes, providing comprehensive thermodynamic insights into molecular interactions.

(Data source: Jiang X, et al. Heliyon. 2024)
Double Electron-Electron Resonance Spectroscopy (DEER)
Double Electron-Electron Resonance Spectroscopy (DEER) is used to study protein structure and interactions within cell lysates or cells and can be used to measure distances within and between molecules. Distance measurement using double electron-electron resonance (DEER, also known as PELDOR) allows distance constraints to be obtained by measuring the dipole interaction between two electron spins. EEER utilizes spin labeling technology to precisely measure intermolecular distances by observing interactions between free radicals and can be applied to complex biological environments. By analyzing signal changes at different delay times, the distance between the spin labels can be inferred.
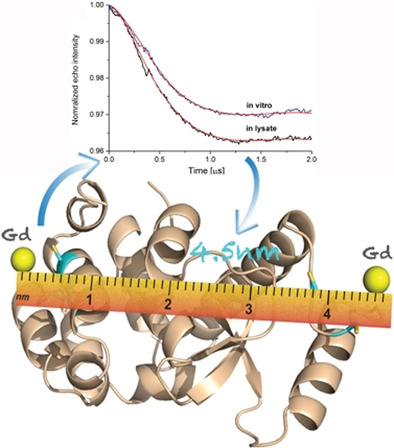
(Data source: Miao Q, et al. Chemistry. 2020)
Proximity labeling (PL)
Proximity labeling (PL) uses engineered enzymes, such as peroxidases or biotin ligases, to genetically tag a protein of interest (POI). PL enzymes convert inert small molecule substrates into short-lived reactive species that diffuse away from the enzyme's active site and covalently label nearby endogenous species. The substrate molecule typically contains a biotin handle, allowing subsequent enrichment of the labeled species using streptavidin beads and identification by mass spectrometry (for proteins) or nucleic acid sequencing (for RNA).
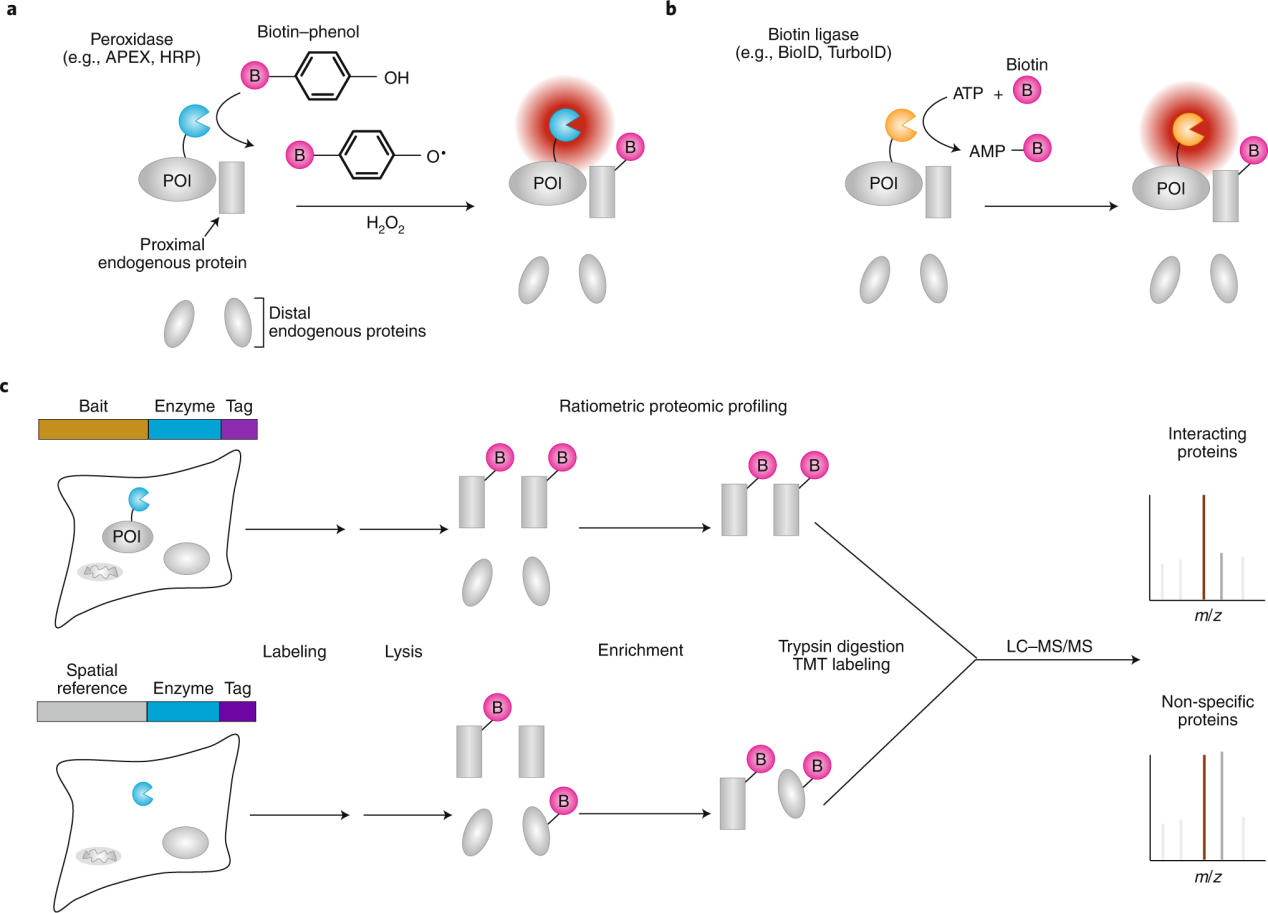
(Data source: Qin W, et al. Nat Methods. 2021)
Comparison of Protein-Protein Interaction Technologies
Each of these protein-protein interaction technologies has its own unique advantages and limitations. When choosing an appropriate method, consider the purpose of the experiment. Combining multiple technologies may provide more comprehensive research results.

