Protein-small molecule interactions play a crucial role in drug development, being central to drug mechanisms of action. With the advancement of medicinal chemistry and molecular biology, experimental techniques for studying small molecule drug-protein interactions have proliferated, providing powerful tools for drug design, drug screening, and understanding drug mechanisms of action. This article will provide an overview of several key methods for studying protein-small molecule interactions, including bio-layer surface interferometry (BLI) , microthermophoresis (MST), small molecule-protein pull-down assays, surface drug affinity reaction targeted stability (DARTS), cell-based thermal mobility shift assay (CETSA), and fluorescence polarization immunoassay (FPIA). Each technique has its unique advantages and application scenarios, and selecting the appropriate experimental method is crucial for uncovering drug-protein interactions.

Biolayer Interferometry (BLI)
Biomembrane interferometry is an optical label-free technique used to monitor interactions between biomolecules in real time, providing data such as binding kinetics and affinity. When visible light passes through a biomembrane layer, it is reflected at the two interfaces of the membrane to form interference waves. The ligand immobilized on the biosensor interacts with the molecules in the analytical solution, causing the thickness of the biomembrane layer to increase, the interference spectrum curve to shift to longer wavelengths, and the phase of the light wave to change. By monitoring this phase change, the interaction between molecules can be accurately measured and the affinity (KD), association rate (ka), and dissociation rate (kd) can be calculated. BLI is a powerful tool for studying interactions between biomolecules such as proteins, antibodies, nucleic acids, polysaccharides, lipids, small molecule drugs, viruses, bacteria, and cells.
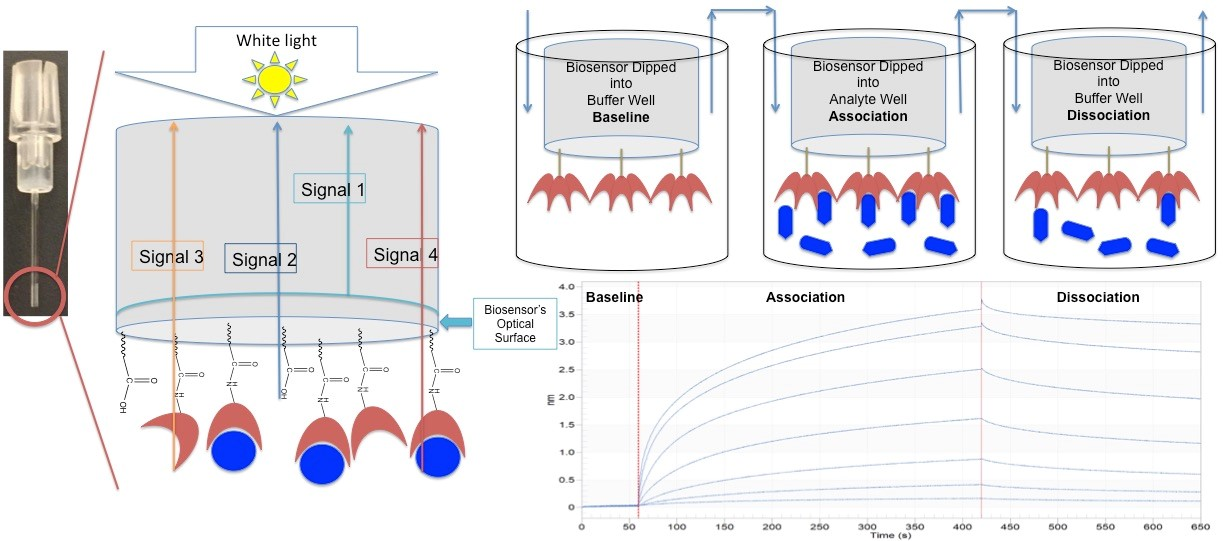
(Data source: RJi Y, et al. Adv Exp Med Biol. 2018)
Microthermophoresis (MST)
Microthermophoresis (MST) is an emerging biomolecular interaction analysis technique. It studies the binding and dissociation processes of biomolecules by monitoring changes in their mobility across a temperature gradient, thereby providing detailed information on these interactions. MST has a wide range of applications in life sciences, including drug screening, signal transduction, molecular detection, and medical diagnostics.
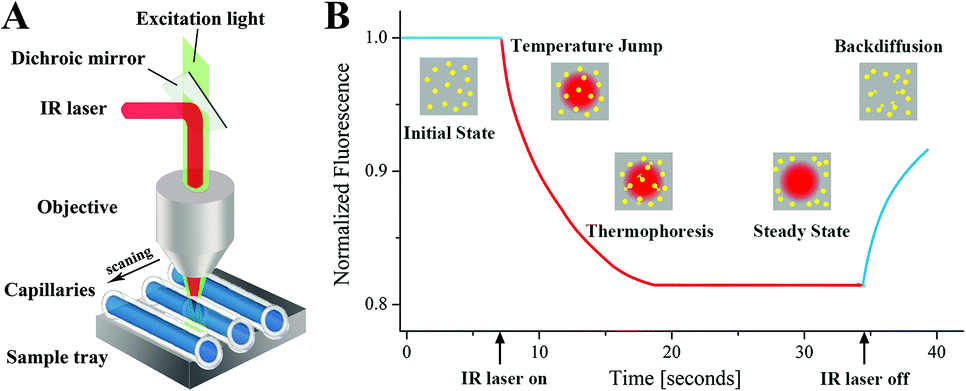
(Data source: Liu Y, et al. Analyst. 2015)
Small molecule-protein pull-down technology
Small molecule-protein pull-down technology uses chemically modified small molecule compounds, such as biotin tags, to screen proteins that interact with small molecules from protein mixtures extracted from cells or tissues using affinity chromatography. Subsequently, these proteins are identified and screened using mass spectrometry to identify the specific binding partners of the small molecule.

(Data source: Smith RJ, et al. STAR Protoc. 2023)
Cell thermal migration assay (CETSA)
The Cellular Thermal Shift Assay (CETSA) is a test used to assess the binding efficiency of drugs to target proteins in cells and tissues. When a target protein binds to a small molecule drug, this binding often enhances the protein's thermal stability. As the temperature gradually increases, unbound protein degrades, while drug-bound protein remains intact due to the drug's protective effect. Consequently, the drug-bound protein exhibits increased stability in its thermal melt curve, with the curve shifting toward higher temperatures (rightward). Detection methods primarily include immunoblotting and protein profiling.

(Data source: Sanchez TW, et al. ACS Chem Biol. 2022)
Fluorescence polarization immunoassay (FPIA)
Fluorescence polarization immunoassay (FPIA) can be used to efficiently and accurately measure the interaction between small molecule drugs and specific target proteins. When a small molecule drug binds to a target protein, the movement of its labeled fluorescent molecules in solution slows down, and this change causes a significant shift in fluorescence polarization. By monitoring this change in fluorescence signal, FPIA can rapidly detect the binding of small molecules to proteins.

(Data source: Liu YOgura Y, et al. Anal Sci. 2023)
Drug-Affinity Response Targeting Stability DARTS
Drug Affinity Response Target Stability (DARTS) is a novel target discovery method that excels at screening small molecule (SM) targets without requiring any structural modification. When a small molecule drug binds to its target protein, it reduces its susceptibility to protease degradation, thereby increasing its stability. The DARTS method can reveal drug-target interactions in cells or tissues by tracking changes in protein stability that act as receptors for bioactive SMs. DARTS can be combined with other techniques, such as CO-IP, SPR, and MST, to identify and validate target proteins.
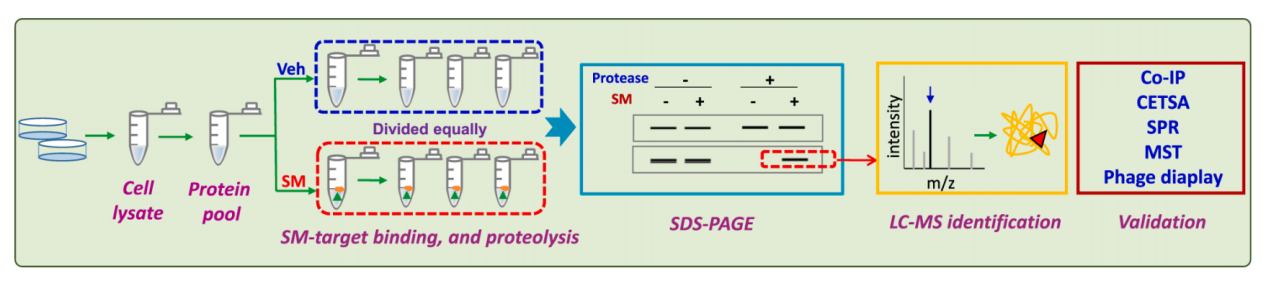
(Data source: Ren YS , et al. Biochem Pharmacol. 2021)
Comparison of small molecule-protein interaction technologies
Each technique has its own unique advantages and limitations, and choosing the right experimental method depends on the research objectives, available resources, and experimental conditions. For example, if you need to study drug effects in cells, CETSA may be a better choice; while for studies requiring high-throughput screening, MST and FPIA may be more appropriate.

