Background
Recombinant expression, isolation, and characterization of perforins are among the most common strategies used to understand their membrane permeability properties. One technique for studying them involves reconstitution into polarized lipid bilayers. Conventional staining methods, such as Coomassie Brilliant Blue, are not sensitive enough to detect all proteins present in a sample. Therefore, obtaining samples that are sufficiently active for reconstitution experiments and pure enough to produce consistent and reproducible results is crucial.
In October 2024, researchers published an article titled " How to isolate channel-forming membrane proteins using the E. coli expression system " in nature protocols. The article describes how to use Escherichia coli BL21Gold (de3) ΔABCF strain to optimize the production of target proteins, and describes in detail how to separate bacterial components by different solubility and how to use detergent solutions to extract target proteins. In addition, the article also introduces how to improve protein purity by ion exchange chromatography and insert purified proteins into outer membrane vesicles for co-purification. After optimization, this protocol can be adapted to the separation of receptors, carriers, pumps or any other membrane-active proteins.

Advantages of E. coli BL21Gold (de3) ΔABCF strain
This strain has been genetically engineered to remove the four major outer membrane porin genes, OmpF, OmpC, LamB, and OmpA, thereby reducing the expression of endogenous membrane proteins. This helps reduce contamination during target protein purification and improves the purity of the target protein.
Expression and purification of channel-forming membrane proteins
Channel-forming membrane proteins fall into three main categories: pore-forming toxins (PTFs), cell wall channel proteins, and outer membrane proteins (OMPs). Regardless of whether the transmembrane domain of PTFs is based on α-helices or β-sheets, they are all soluble in detergent-free buffers. When the amount of inducer, length of expression, and temperature are optimized, PTFs can be collected in the supernatant after the initial ultracentrifugation. Cell wall channel proteins, such as those of the MspA family, solubilize in a similar manner within a short time; after extraction, they should be purified from soluble fractions such as PFTs, but the purification buffer must contain some detergent (e.g., 0.5% octyl-POE). Most OMPs and membrane proteins typically end up in inclusion bodies and must be extracted from these inclusion bodies by resuspending the appropriate pellet with an appropriate detergent and at an appropriate temperature for a period of time. Once the protein displays a clear band in the supernatant, it can be purified using a chromatography step.

If the protein of interest is a channel-forming membrane protein natively expressed in the outer membrane of Gram-negative bacteria, its activity is likely to be affected by the surrounding lipopolysaccharide (LPS). Fusion of OMVs into a planar bilayer structure allows for the characterization of recombinant porins in their native environment. When copurifying a protein with OMVs, several important details must be considered: first, the protein must be translated with a signal peptide compatible with the E. coli machinery; second, protein expression must be induced for 24 hours to stimulate the production of OMVs; and the strain must be deficient in OmpA, a characteristic of OMV overproduction. This results in increased product yields.
Membrane channel protein expression and purification issues and optimization strategies
The authors encountered a series of problems in the process of expressing and purifying membrane channel proteins, and also proposed corresponding optimization strategies to solve these problems.
OD600 of induced expression: When optimizing protein expression conditions, it is recommended to try different target OD600 values, with OD600 being a good starting point. At this starting point, two situations were found after induction at 30°C with a standard amount of inducer:

Case 1: OD600 continues to increase (e.g., OmpE36). This change indicates that protein expression is harmless to bacterial growth.
Case 2: OD600 decreases, levels off, and then increases again (e.g., OprO/P). This indicates that protein expression is detrimental to bacterial growth. In this case, adjust the OD600 induction starting point, inducing for a longer time at a low OD value or a shorter time at a high OD value. Also, lower the induction dose and temperature (20°C).

Expression plasmid: If the protein is not expressed, there may be issues with the plasmid or ORF. Reduce the inducer, induce expression at a lower temperature for a longer period of time, or resequence the plasmid. Codon optimization can improve protein expression. If the expressed protein is toxic to the host, such as when pLysS expresses lysozyme, which inactivates T7 polymerase, increase the inducer (IPTG up to 2 mM) and shorten the expression time at a higher OD.
Expression temperature: Before induction, the culture can be cooled on ice for 15 min. For some long-expressing or highly expressed proteins, it is useful to lower the expression temperature (not less than 20°C) to obtain better folding and less integration into inclusion bodies in favor of the outer membrane.
Protein extraction optimization strategies: Select the appropriate detergent and concentration to aid in the extraction of the target protein from the membrane fraction. Adjust the detergent concentration and extraction time to optimize protein solubility and recovery. Adjust the pH and ionic strength of the extraction buffer to improve the solubility and stability of the target protein.

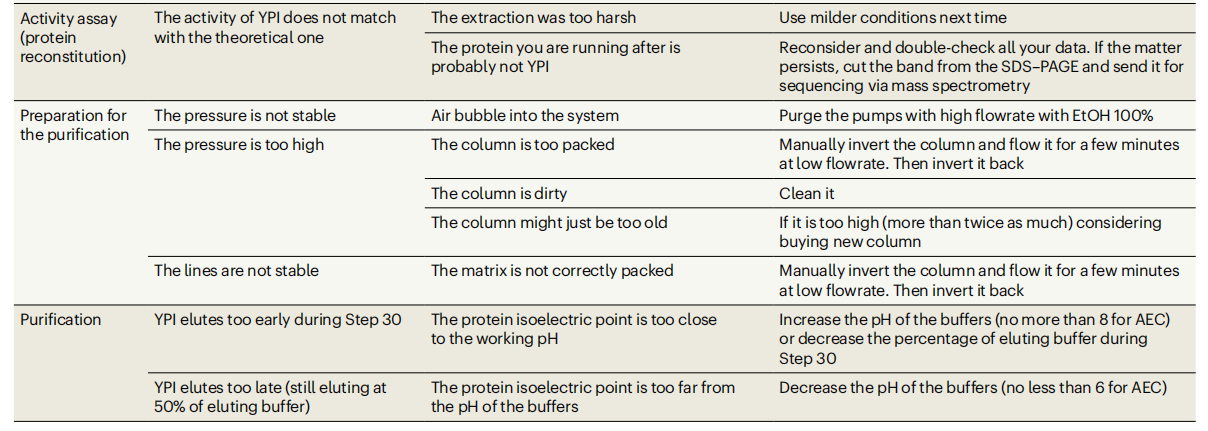
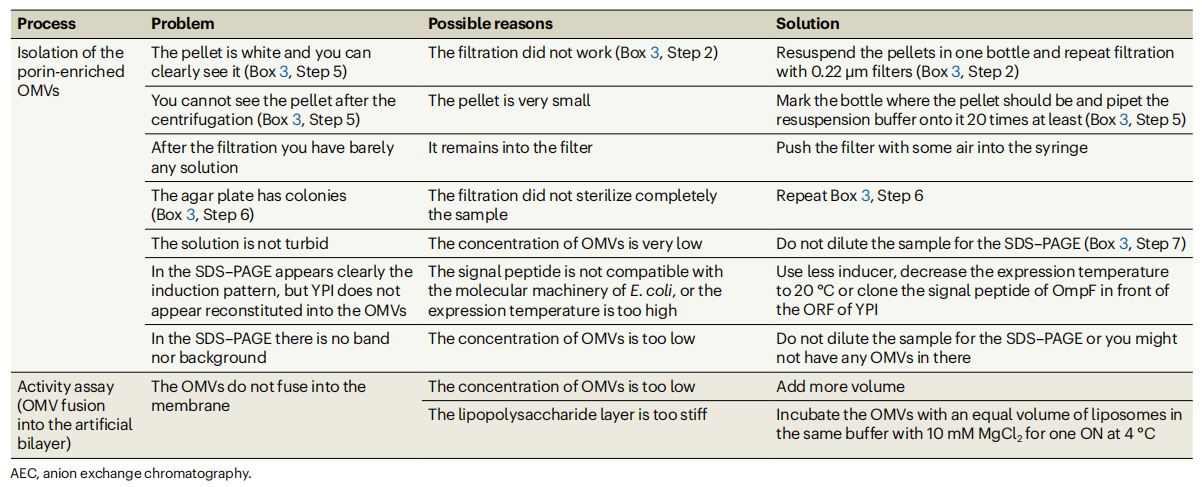
Summarize
The protocol described in this article provides a rational strategy for the step-by-step optimization of the expression, extraction, and purification conditions of channel-forming membrane proteins in the E. coli recombinant system. Expression time and temperature, inducer concentration, detergent properties and concentration, incubation time, and purification pH are all parameters that can be optimized according to the specific protein. By selecting the appropriate induction temperature and induction time, as well as the appropriate detergent, it will be easier to extract the protein in the future. The protocol mentioned in the article is not only applicable to prokaryotic channel-forming membrane proteins, but can also be used for the production of pore-forming proteins in chloroplasts, mitochondria, or general eukaryotic organisms. Through proper optimization, this protocol can also be adapted to the isolation of receptors, carriers, pumps, or any other membrane-active proteins, thus providing a powerful tool for a wide range of biomedical research.
