Background
Natural proteins are highly optimized for function but are often difficult to produce at scales suitable for biotechnological applications due to poor expression in heterologous systems, limited solubility, and temperature sensitivity. Therefore, a general approach to improve the physical properties of natural proteins while maintaining their functionality is needed, which can be broadly applied to protein-based technologies.

On January 9, 2024, researchers from the Institute for Protein Design at the University of Washington published a study titled "Improving Protein Expression, Stability, and Function with ProteinMPNN" in the Journal of the American Chemical Society. The study demonstrated that the deep neural network ProteinMPNN tool provides a path to increase protein expression, stability, and function. For myoglobin and tobacco etch virus (TEV) protease, designs with improved expression, increased melting temperature, and improved function were generated. For TEV protease, multiple designs with higher catalytic activity compared to the parent sequence and previously reported TEV variants were identified. This approach should have broad application in the future to improve the expression, stability, and function of important biotechnological proteins.

Protein stabilization using ProteinMPNN
The design space was chosen to preserve the protein's native function by fixing the amino acid properties of residues close to the ligand and those that are highly conserved in multiple sequence alignments. The protein backbone structure and fixed position information were input into ProteinMPNN to generate novel amino acid sequences that could fold into the input structure. The backbone structure of the loop region can optionally be reshaped using RoseTTAfold joint mapping to further idealize the input protein.

Design of myoglobin variants with improved stability
The authors applied the above-described ProteinMPNN design approach using the crystal structure of human myoglobin nMb. Seventeen positions around the heme ligand in the heme-bound structure were fixed to maintain the oxygen-binding function of myoglobin. Sixty sequences were generated using ProteinMPNN and evaluated for their potential to reproduce the myoglobin backbone coordinates using AlphaFold2 single sequence prediction. The authors also explored limited backbone redesign of less ordered regions to further stabilize the protein. These less conserved loop regions were selected and the RoseTTAFold technique was used for backbone remodeling, generating two different sets of structural remodeling designs: one set redesigned the region connecting helices E and F, and the other set additionally included the CD loop region. From these remodeled backbones, sequence design was again performed using ProteinMPNN, with the heme-binding site remaining unchanged as described above.
Synthetic genes encoding the designed and parental sequences were synthesized and expressed in Escherichia coli. Thirteen of the 20 designs (up to 4.1-fold) achieved higher total soluble protein yields than native myoglobin. Eight designs had higher melting temperatures than native myoglobin, and six of these remained fully folded at 95°C (native myoglobin melts at 80°C). dnMb19, generated using a more aggressive backbone remodeling strategy, exhibited significantly greater thermal stability for heme binding compared to native myoglobin. RoseTTAFold, combined with the power of mapping technology and ProteinMPNN, accurately remodeled the native protein backbone, simultaneously improving solubility, thermal stability, and functional stability.

Design of TEV protease variants with improved stability and catalytic activity
By applying this design strategy to the tobacco etch virus (TEV) cysteine protease, an enzyme used to specifically remove purification tags from recombinant proteins, the utility of ProteinMPNN sequence design for enzyme stabilization was explored. Four different designs were generated, fixing only the amino acid identities of active site residues and residues that are highly conserved within the TEV protease family. Backbone redesign was then performed in regions less conserved within the TEV protease family.
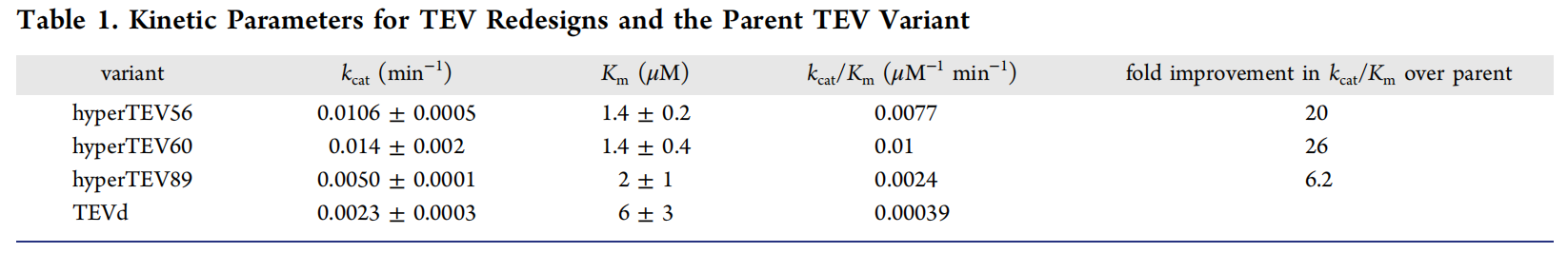
A total of 144 sequences were generated using ProteinMPNN. 134 of the 144 designs were expressed in a soluble form and eluted as monomers by SEC. 129 of the 144 designs exhibited higher soluble expression levels than TEVd. 64 designs exhibited substrate peptide cleavage activity, and designs that did not fix conserved residues had increased expression levels but lacked functional activity. Enzyme catalytic activity assays revealed that hyperTEV60 retained 90% of its original cleavage activity, while TEVd was reduced to 15% of its original activity, indicating that the stability of the designed TVEs was significantly improved.

Microsecond molecular dynamics (MD) simulations of the TEV-peptide complex were performed to investigate the effects of the introduced mutations on overall protein dynamics. Compared to TEVd, widespread rigidification of loop regions distributed throughout the structure was observed in the designed variants. This backbone rigidification in distal regions not directly involved in substrate binding may be associated with allosteric improvements in substrate binding, as reflected by a two- to three-fold decrease in the K m values measured for the designed variants.
Summarize
This study demonstrates that ProteinMPNN can be used to improve the expression, stability, and function of native proteins, guided by existing sequence and structural information. While the optimal number of residues to maintain (or perhaps enhance) function may have to be determined empirically in each case, this procedure is straightforward, facilitated by ProteinMPNN's computational efficiency and ease of use, and requires testing far fewer variants than typical experimental screens. This technology may have broad future applications in protein expression.
