TurboID is an engineered biotin ligase that uses ATP to convert biotin into biotin-AMP, an active intermediate that covalently labels neighboring proteins. Optimized through directed evolution, TurboID exhibits significantly higher activity than previously described biotin ligase-based proximity labeling methods, such as BioID, enabling higher temporal resolution and broader applications. Split-TurboID consists of two inactive fragments of TurboID that can reassemble through protein-protein or organelle-organelle interactions, contributing to higher targeting specificity than the full-length enzyme alone. Proteins biotinylated by TurboID or split-TurboID are subsequently enriched with streptavidin beads and identified by mass spectrometry.

In November 2020, researchers published an article titled "Proximity labeling in mammalian cells with TurboID and split-TurboID" in Nat Protoc, describing a protocol for proximity labeling in mammalian cells using TurboID and split-TurboID. The authors outline how to use the lab's latest PL enzymes, TurboID6 and split-TurboID, for subcellular proteomic mapping. They describe in detail how to design and characterize fusion constructs (variable time), prepare proteomic samples (5-7 days), acquire mass spectrometry data (2 days), and analyze proteomic data (1 week).

Limitations of TurboID Proximity Tagging Technology
Different subcellular compartments have varying pH, redox environments, and endogenous nucleotide concentrations, leading to variations in TurboID activity. TurboID activity also varies across organelles and organisms; and its fusion to proteins may affect protein stability, localization, and function. Therefore, evaluating the activity of TurboID fusion constructs intended for proteomics is crucial. Such assays may not account for dually localized proteins, resulting in reduced coverage (for example, a protein present in both the ERM and the cytoplasm would be eliminated in a second round of screening using the cytoplasmic TurboID as a reference).
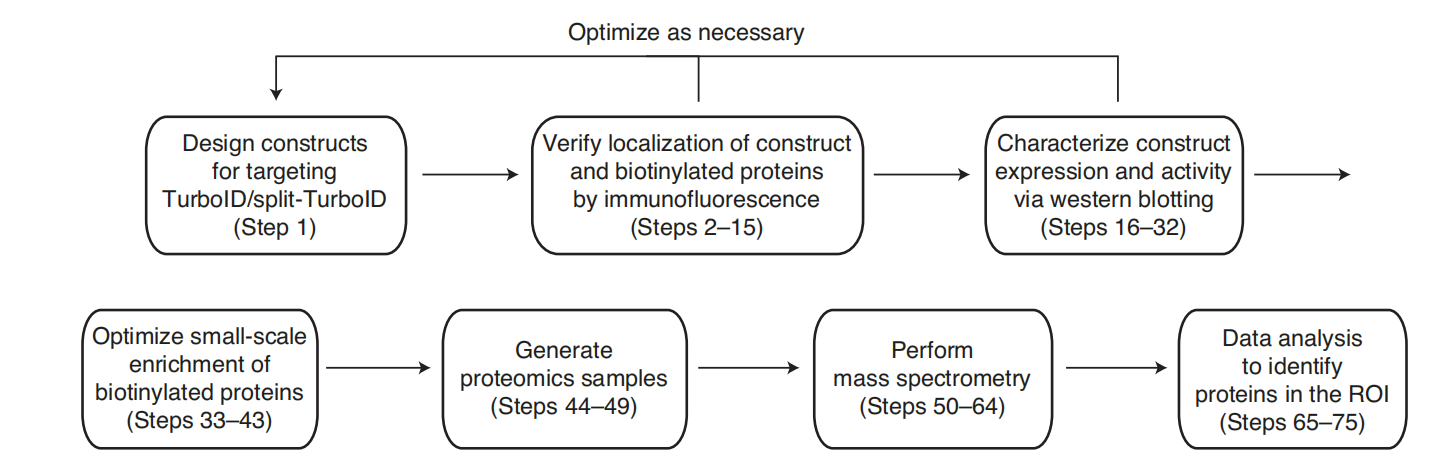
Overview of the TurboID proteomics experiment workflow
The workflow for performing a TurboID proteomics experiment is as follows: the designed fusion constructs should be characterized using immunofluorescence and western blotting and optimized if necessary. Following construct validation, enrichment conditions should be optimized before generating proteome samples for mass spectrometry analysis. Finally, data analysis is performed after mass spectrometry analysis to identify enriched proteins. Data analysis primarily uses ratiometric methods combined with receiver operating characteristic (ROC)-based threshold analysis.
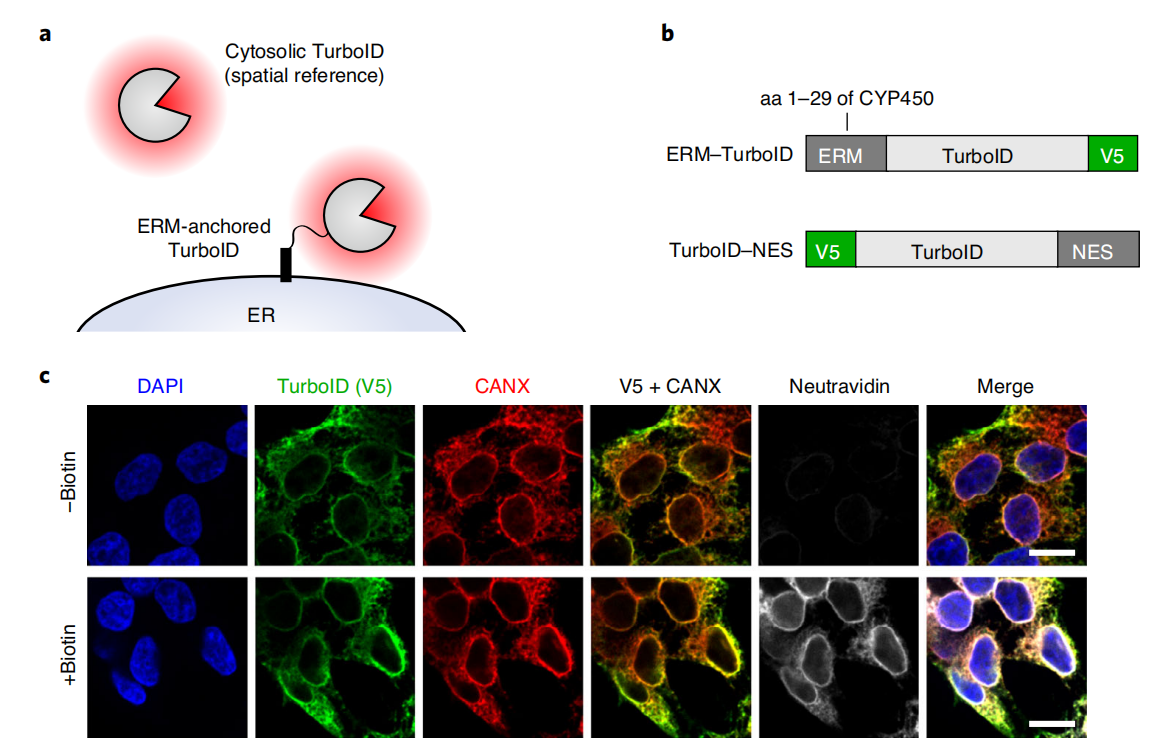
Position TurboID in the target area
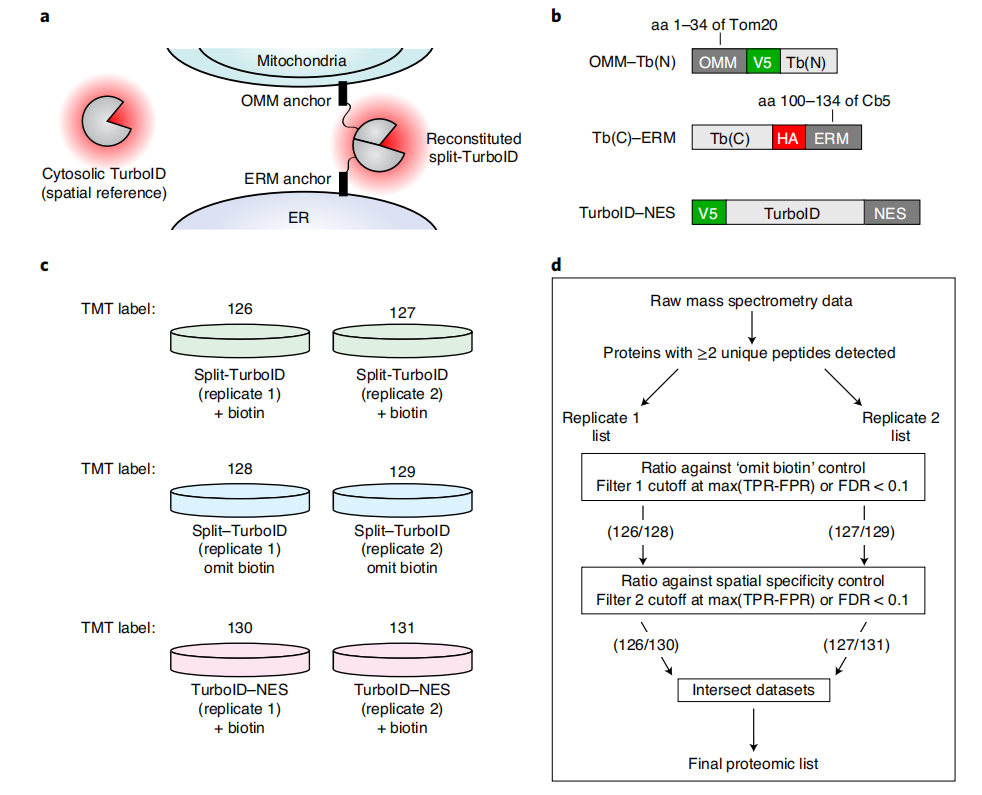
First, the construct must be targeted to the cellular region or protein complex of interest . For proteomic mapping of organelles or subcellular structures, targeting can be achieved through the use of localization sequences. TurboID and split-TurboID are targeted to the extracellular matrix (ERM) using targeting peptides derived from cytochrome P450 and cytochrome b5, and to the outer membrane (OMM) using targeting peptides derived from mitochondrial antiviral signaling protein (MAVS) and outer membrane 20-kilodalton subunit translocase (Tom20). Construct localization can be verified by immunostaining and comparison with co-transfected/infected fluorescent protein markers targeting the same organelles, or by co-immunostaining with endogenous markers of the targeted region.
Notes:
1)Avoid targeting the target region by fusion with the full-length functional protein, as overexpression of the bioactive protein may interfere with signal transduction and biological function.
2)After fusion, there may be unique subcellular localization, and multiple fusion constructs need to be tested at different levels to ensure correct localization and minimize interference with the target.
3)TurboID or split- TurboID fragments are fused directly to the bait protein under study. It is critical to ensure that the fusion of TurboID/split- TurboID fragments does not perturb the localization pattern, biological function, or interactions of the bait protein.
4)To ensure that TurboID and split- TurboID do not affect ligase activity.
5) If the construct does not colocalize with the targeted region or if extensive organelle perturbation is observed, construct optimization is necessary: modify the linker length or rigidity, construct geometry, and targeting sequence. Glycine- and serine-rich linkers are generally used, starting at 10 amino acids. Avoid FLAG and other lysine-rich epitope tags, which may be destroyed by TurboID labeling chemistry. TurboID/split-TurboID fusion constructs can be introduced into mammalian cells via transfection or viral transduction.
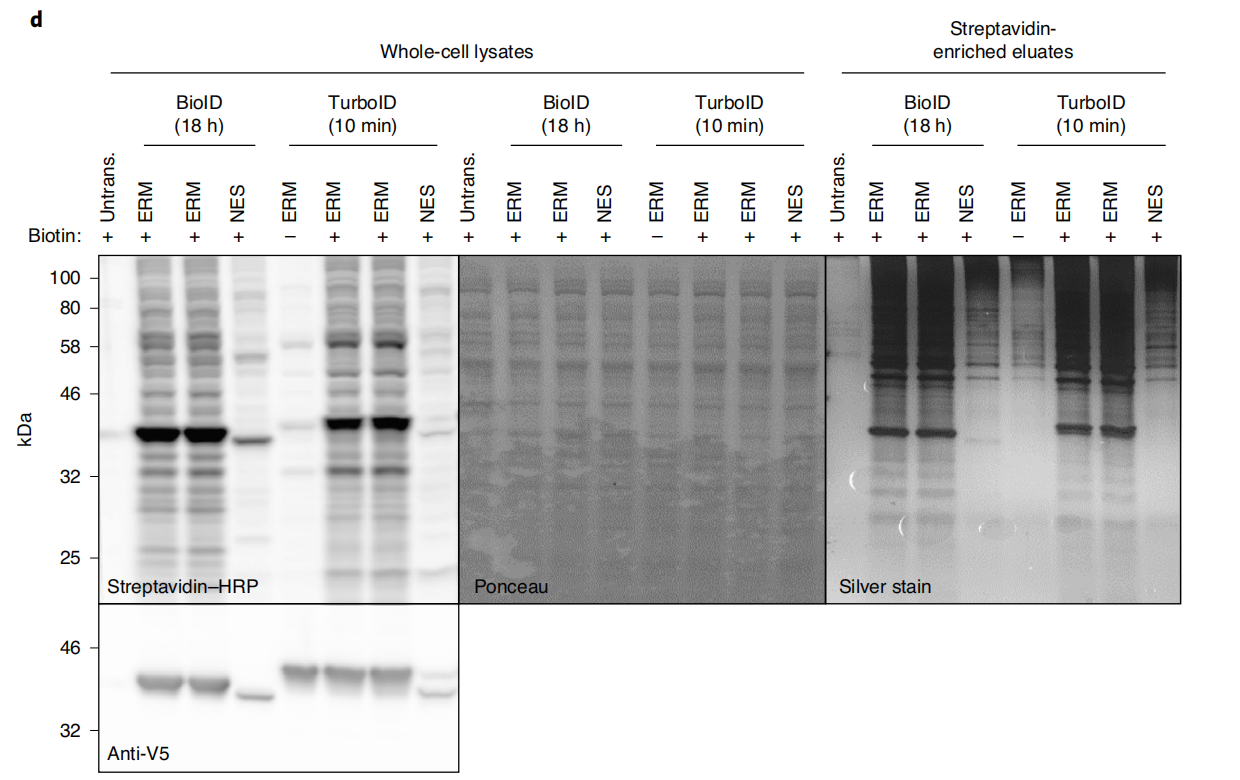
Characterization of the activity of TurboID constructs
After verifying the correct localization and/or functionality of the fusion construct, TurboID/split-TurboID activity should be assessed. This can be accomplished by immunofluorescence staining of biotinylated proteins (e.g., using a fluorescein-avidin conjugate) or by Western blot analysis of biotinylated proteins in whole-cell lysates (e.g., using streptavidin-fluorescein or streptavidin-HRP conjugates). The same lysates should also be run and blotted using a fluorescein/HRP antibody conjugate against the corresponding epitope tag to verify the expression and integrity (lack of proteolytic enzymes) of the TurboID fusion construct. Before large-scale proteomic experiments, perform a small-scale enrichment of TurboID/split-TurboID biotinylated proteins. Analyze the eluate samples by silver staining to confirm successful enrichment of total protein in the experimental samples and compare to a negative control.
Notes:
1)Include negative controls without ligase or biotin to assess signal and background biotinylation levels of endogenously biotinylated proteins;
2)Use the lowest biotin concentration and shortest labeling time to avoid nonspecific labeling;
3)Optimize various experimental conditions, such as changing the volume of magnetic beads and the number of washes, to maximize the capture of biotinylated species while minimizing nonspecific binding to the beads;
4)Before enrichment, free biotin can be removed using a gel filtration column. Depletion of endogenous biotinylated proteins before enrichment can increase the signal of TurboID/split-TurboID labeled proteins.
Proteomics sample preparation
Conditions optimized from small-scale enrichment can be scaled up for large-scale proteomics experiments. For proteomics experiments in HEK293T cells, vials of approximately 20 million cells are used to generate samples for mass spectrometry. Enzymatic digestion of streptavidin beads with trypsin or other hydrolytic enzymes releases proteins bound to the affinity matrix. After removal of the streptavidin beads, the released proteins are completely digested into peptides for analysis by liquid chromatography - tandem mass spectrometry (LC - MS/MS). A variety of quantitative and semiquantitative methods can be used to distinguish between protein enrichment under experimental conditions and negative controls. The quantification method used in this experiment was translocation mass spectrometry tag (TMT).

Note: Duplicate samples should be included for each experimental condition, including negative controls without ligase or biotin, as well as spatially specific controls. For example, in the analysis of ERM proteins using TurboID or the analysis of ER-mitochondria contact proteins using split-TurboID, samples containing TurboID-NES localized to the cytosol should also be included for comparison.
Data Analysis
For quantitative methods, two ROC-based screening steps are required: first, identifying biotinylated proteins; and then determining which proteins are preferentially labeled by TurboID targeting the region of interest.
The displayed histogram of the distribution of true-positive and false-positive proteins in the dataset, plotted by TMT ratio, demonstrates successful enrichment of true-positive proteins using the ROC-determined cutoff. Proteins with a TMT ratio above the determined cutoff are considered biotinylated, while proteins below the cutoff are considered likely nonspecific binders. A secondary screen aims to compare experimental samples with a TurboID reference control targeting overlapping but distinct subcellular regions (i.e., located in adjacent or contiguous subcellular regions). This step also involves calculating the TMT ratio and analyzing the ROC cutoff to determine the TMT signal of the experimental sample with that of a reference sample (e.g., cytoplasmic TurboID-NES). Proteins are further filtered to determine which proteins are preferentially biotinylated in the target region.
Note: These 2 filtering steps need to be repeated and performed independently for each sample, and the final proteome list is generated from the intersection of the filtered lists of each replicate sample.
Common Problems and Solutions
The authors also summarize the problems that may arise when using proximity labeling technology and their corresponding solutions.
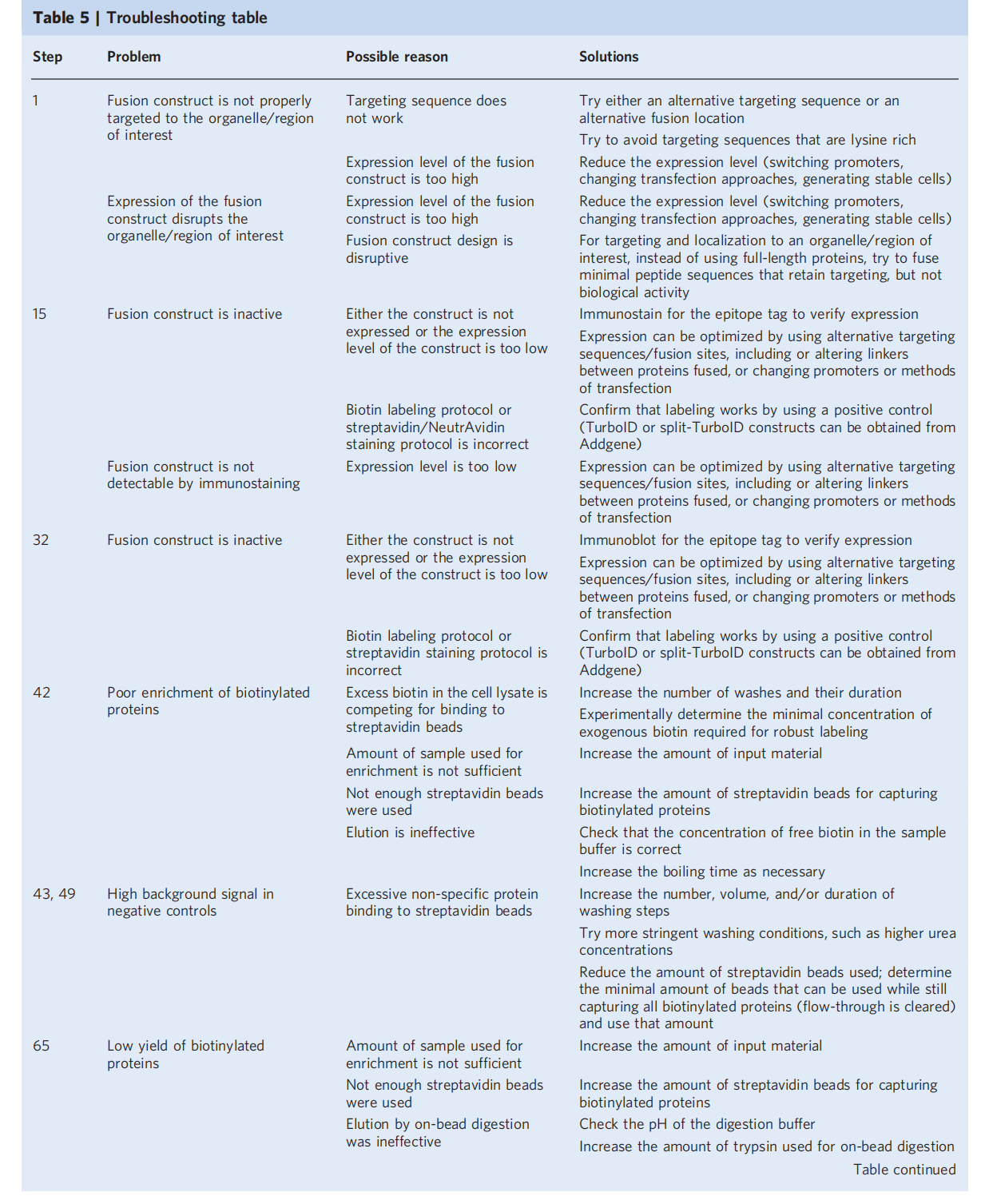
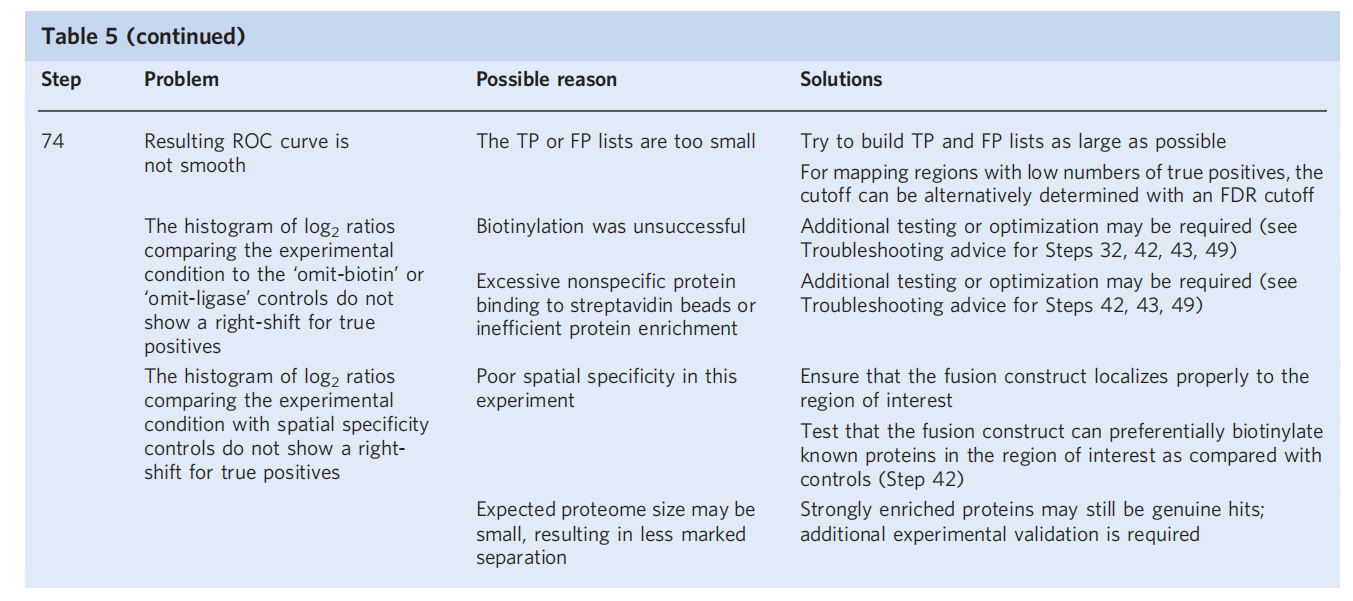
Expected experimental results
Targeted TurboID or segmented TurboID fusion constructs should colocalize with appropriate markers. For a successful proteomics experiment, TP proteins should have higher TMT ratios than FP proteins, and thus the receiver operating characteristic (ROC) plot should show that the TPR increases faster than the FPR. Plotting the distribution of TP and FP proteins should also demonstrate that the determined TMT ratio threshold successfully enriches for TP proteins. For example, in proteomics mapping of ERMs, using a threshold defined by the maximum TPR-FPR, ERM proteins are significantly enriched relative to cytosolic proteins. For mapping regions with known low true positives, such as ER-mitochondria contact sites, thresholds can be determined using an FDR threshold. Candidate proteins generated by proteomics experiments should also be validated using independent methods, such as immunoblotting, pull-down assays, and functional assays, to verify that their protein interaction results are genuine.
Summarize
This article provides a detailed experimental protocol for using TurboID and split-TurboID technologies for proximity labeling in mammalian cells to investigate protein-protein interactions and subcellular proteomic profiling. Through optimized experimental design and rigorous data analysis, this technology enables high-resolution visualization of molecular interactions within living cells. The article also discusses potential limitations and precautions encountered during the experiment to ensure reproducibility of the method and reliability of the results. This work not only advances biomedical research but also provides new tools for exploring disease mechanisms and developing new drugs.
